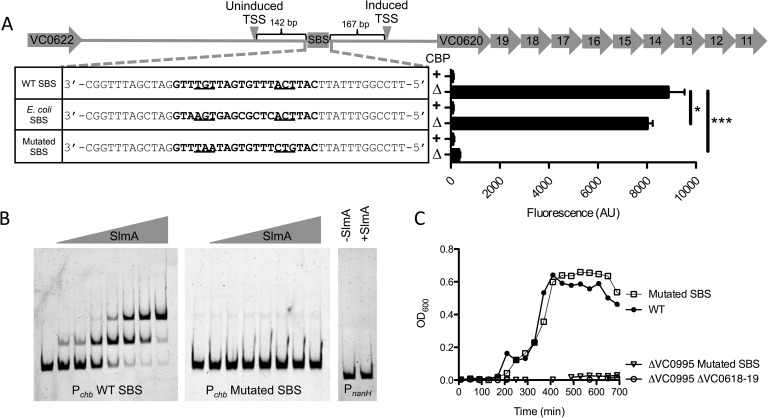
Fig 2. SlmA is a direct transcriptional activator of Pchb.
(A) Promoter map of Pchb showing the SBS and the TSSs mapped in the presence (induced) and absence (uninduced) of chitin. “WT SBS” represents the native Pchb sequence, “E. coli SBS” indicates that the native SBS was swapped for the consensus SBS from E. coli, and “mutated SBS” indicates that the 6 highly conserved residues in the SBS were mutated. SBS sequences for the indicated mutants are shown in bold text and the 6 most highly conserved residues of the SBS are underlined. GFP fluorescence was measured in strains harboring the indicated mutations in the Pchb-gfp transcriptional reporter. The cbp status of each strain is indicated by a “+” (cbp intact) or a “Δ” (Δcbp strain background). Data are shown as the mean ± SD and are from at least three independent biological replicates. (B) EMSAs performed with purified SlmA and the indicated promoter probes. The two Pchb probes were incubated with (from left to right) 0 nM, 7.5 nM, 15 nM, 30 nM, 60 nM, 120 nM, 240 nM, or 480 nM SlmA. The PnanH probe was incubated with 0 nM or 480 nM SlmA. (C) Growth curves of the indicated strains in M9 minimal medium containing 0.5% chitobiose as the sole carbon source. Data from B and C are representative of at least 2 independent experiments. *** = p<0.001, * = p<0.05, NS = not significant.