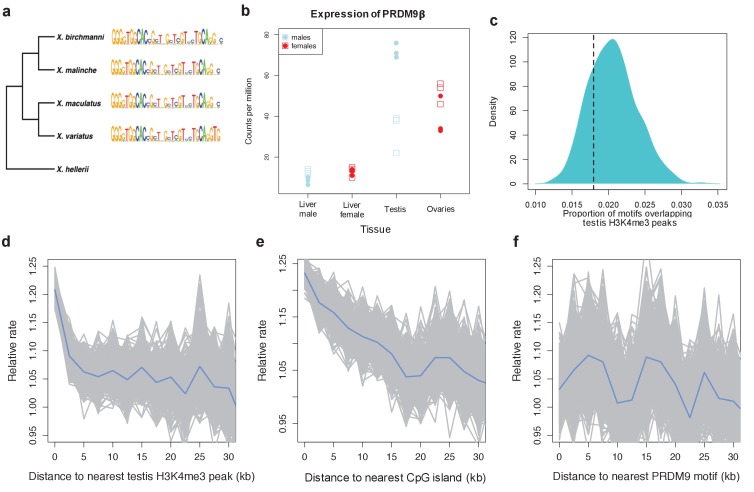
Figure 4. Patterns of recombination and PRDM9 evolution in swordtail fish.
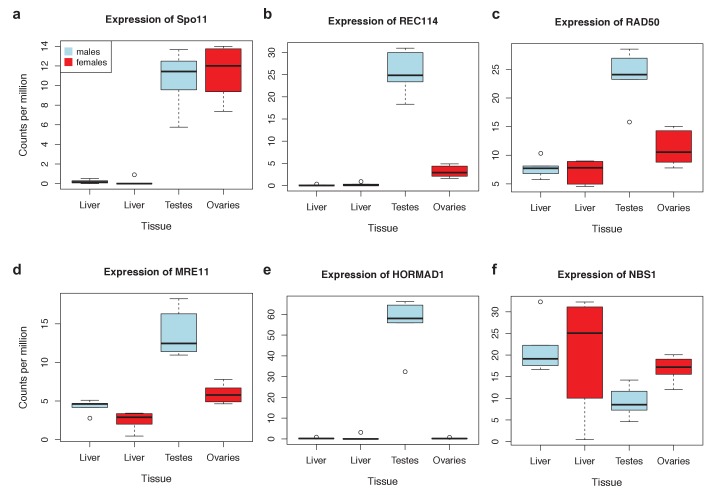
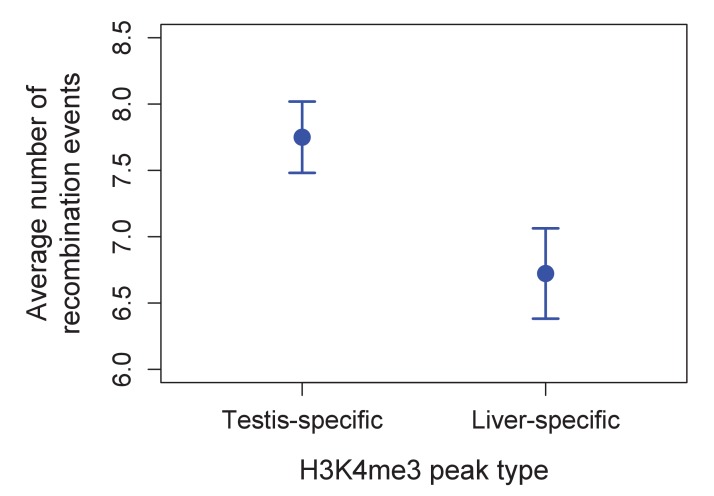
(a) The ZF array of PRDM9 appears to be evolving slowly in Xiphophorus, with few changes over 1 million years of divergence (Cui et al., 2013; Jones et al., 2013). (b) PRDM9 is upregulated in the germline relative to the liver in X. birchmanni (circles) and X. malinche (squares; panel shows three biological replicates for each species). (c) The computationally-predicted PRDM9 binding sites is not unusually associated with H3K4me3 peaks in testes (d) Crossover rates increase near H3K4me3 peaks in testis (e) Crossover rates increase near CGIs (f) Crossover rates do not increase near computationally-predicted PRDM9 binding sites (see Figure 4—figure supplement 3 for comparison). Crossover rates were estimated from ancestry switchpoints in naturally occurring hybrids between X. birchmanni and X. malinche (see Materials and methods).