Abstract
E1 enzymes for ubiquitin (Ub) and Ub-like modifiers (Ubls) harbor two catalytic activities that are required for Ub/Ubl activation: adenylation and thioester bond formation. Structural studies of the E1 for the Ubl small ubiquitin-like modifier (SUMO) revealed a single active site that is transformed by a conformational switch that toggles its competency for catalysis of these two distinct chemical reactions. Although the mechanisms of adenylation and thioester bond formation revealed by SUMO E1 structures are thought to be conserved in Ub E1, there is currently a lack of structural data supporting this hypothesis. Here, we present a structure of Schizosaccharomyces pombe Uba1 in which the second catalytic cysteine half-domain (SCCH domain) harboring the catalytic cysteine has undergone a 106° rotation that results in a completely different network of intramolecular interactions between the SCCH and adenylation domains and translocation of the catalytic cysteine 12 Å closer to the Ub C terminus compared with previous Uba1 structures. SCCH domain alternation is accompanied by conformational changes within the Uba1 adenylation domains that effectively disassemble the adenylation active site. Importantly, the structural and biochemical data suggest that domain alternation and remodeling of the adenylation active site are interconnected and are intrinsic structural features of Uba1 and that the overall structural basis for adenylation and thioester bond formation exhibited by SUMO E1 is indeed conserved in Ub E1. Finally, the mechanistic insights provided by the novel conformational snapshot of Uba1 presented in this study may guide efforts to develop small molecule inhibitors of this critically important enzyme that is an active target for anticancer therapeutics.
Keywords: conformational change, enzyme mechanism, enzyme structure, structure-function, ubiquitin, X-ray crystallography, E1, adenylation, domain alternation, thioester
Introduction
Ubiquitin (Ub)2 and ubiquitin-like modifiers (Ubls) play a fundamental role in almost every aspect of eukaryotic biology through their regulation of myriad cellular proteins (1–3). E1 enzymes function as the gatekeepers of Ub/Ubl conjugation cascades by specifically binding and activating their cognate Ub/Ubl in a two-step ATP·Mg-dependent process. Ub/Ubl activation involves adenylation of the Ub/Ubl C terminus in the first step followed by nucleophilic attack of the acyladenylate by the E1 catalytic cysteine to form a high energy E1∼Ub thioester intermediate in the second step (4, 5). E1 next recruits E2 conjugating enzymes and transfers Ub/Ubl to the E2 catalytic cysteine in a process termed E1-E2 thioester transfer (or transthiolation) (6–8). The resulting E2∼Ub thioester intermediate then functions with a wide array of E3 ligases that define substrate specificity and catalyze Ub/Ubl conjugation to target proteins through a variety of mechanisms (9–13).
So-called “canonical” Ub/Ubl E1 enzymes, such as those for the Ub, SUMO, and NEDD8 conjugation cascades, are defined by a conserved multidomain structure in which each domain plays a distinct role in their catalytic cycle (14–17). Active and inactive adenylation domains (AAD and IAD, respectively) specifically recognize and adenylate the C terminus of their cognate Ub/Ubl prior to thioester bond formation (14–17). The Cys domain, which is split into first and second catalytic cysteine half-domains (FCCH and SCCH domains, respectively), harbors the catalytic cysteine residue that forms the thioester bond with Ub/Ubl (14–17). Finally, the ubiquitin-fold domain (UFD) of a canonical E1 is involved in molecular recognition of its cognate E2(s), which is followed by E1-E2 thioester transfer and formation of the E2∼Ub thioester intermediate (17–20).
In all structures of canonical E1s determined with their respective cognate Ub/Ubl noncovalently bound to the adenylation domains, the SCCH domain adopts an “open” conformation in which the catalytic cysteine that serves as the nucleophile during thioester bond formation is located more than 30 Å away from C terminus of the Ub/Ubl (14–16, 18–22). Recent structural and biochemical studies revealed that SUMO E1 has a single active site that is reconfigured for catalysis of adenylation or thioester bond formation via a series of complementary conformational changes in several regions of the E1 (21). Following adenylation of SUMO, contacts to ATP·Mg are released, facilitating a 130° closure of the SUMO E1 SCCH domain (termed domain alternation) and remodeling of several other structural elements that comprise the catalytic machinery of the adenylation active site (21). As a result, more than half of the residues that promote adenylation are replaced with residues that promote thioester bond formation, and the E1 catalytic cysteine is brought into proximity of the SUMO C terminus (21). The concomitant disassembly of the adenylation active site and assembly of the thioester bond formation active site thereby toggles the catalytic competency of the active site, serving as a mechanism to drive the SUMO activation process forward. Although other canonical E1 enzymes such as Ub E1 (Uba1) and NEDD8 E1 are predicted to undergo similar conformational changes during activation of their respective Ub/Ubl (21, 23, 24), there is currently a lack of structural data proving this hypothesis.
In this study, we present the first crystallographic snapshot of Uba1 in which its SCCH domain has undergone a domain alternation. Comparison of our structure with previously determined Uba1 structures reveals a ∼106° rotation of the SCCH domain and remodeling of several elements within the adenylation active site that are crucial for catalysis. Comparative structural analysis reveals that the observed conformational changes in Uba1 are analogous to those required for adenylation and thioester bond catalysis by SUMO E1. Altogether, our structural and biochemical data suggest that domain alternation and remodeling of the adenylation active site are interconnected and intrinsic structural features of Uba1 and that the structural basis for adenylation and thioester bond formation involving SCCH domain alternation and active site remodeling exhibited by SUMO E1 is indeed conserved in Uba1.
Results and discussion
Structure determination and characterization of a Schizosaccharomyces pombe Uba1/NSC624206 co-complex
NSC624206 is a Ub E1 inhibitor that specifically inhibits thioester bond formation between Ub and Uba1 without affecting adenylation activity (25), and the initial goal of this study was to elucidate the molecular mechanism by which NSC624206 inhibits Ub E1 activity. Given the disulfide bond present within NSC624206, Ungermannova et al. (25) concluded that the mechanism of inhibition is through covalent attachment of one “fragment” of NSC624206 to the Uba1 catalytic cysteine via disulfide bond, thereby abolishing its reactivity, with the other fragment serving as a leaving group during adduct formation. Thus, we used a Uba1 construct harboring a catalytic cysteine to alanine substitution (C593A) in our crystallographic studies to prevent adduct formation between Uba1 and NSC624206, thereby allowing us to potentially identify the Uba1-binding site for intact inhibitor.
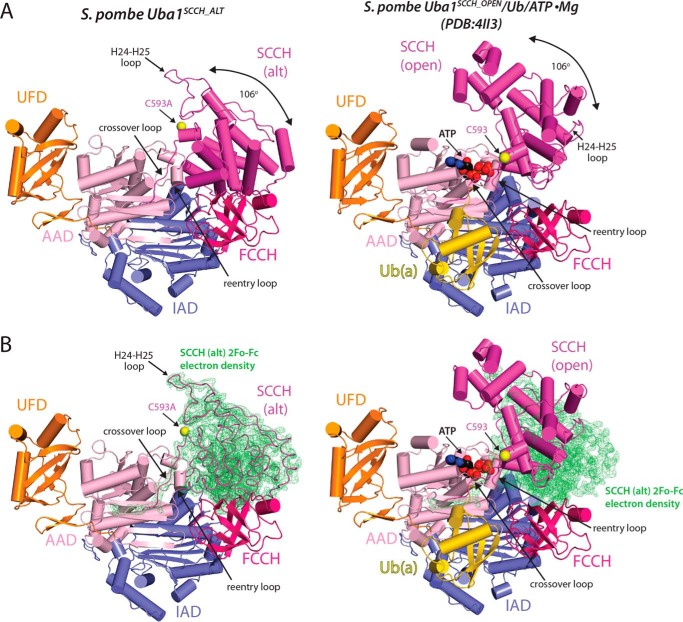
We obtained diffracting crystals of an S. pombe Uba1/NSC624206 co-complex in a crystal form distinct from that of any published Uba1 structure and collected a complete data set to 2.79 Å (Table 1). Initial efforts to solve the structure by molecular replacement using existing Uba1 structures as the search models failed. Deletion of the SCCH domain from the search model yielded a molecular replacement solution, and inspection of the resulting maps revealed clear electron density for the SCCH domain in a drastically different conformation compared with other Uba1 structures (Fig. 1, A and B). A subsequent multidomain molecular replacement approach resulted in successful placement of the SCCH domain (Fig. 1B), and the resulting structure (hereafter referred to as Uba1SCCH_ALT) was refined to R/Rfree values of 0.206/0.240 (Table 1).
Table 1.
Crystallographic data and refinement statistics
Parentheses indicate statistics for the high-resolution data bin for x-ray data. APS, Advanced Photon Source. CC1/2 is the correlation coefficient between two random half data sets.
| S. pombe Uba1 | |
|---|---|
| Data collection | |
| Protein Data Bank code | 5UM6 |
| Source | APS 22 ID |
| Wavelength (Å) | 1.08 |
| Resolution limits (Å) | 50–2.79 (2.90–2.79) |
| Space group | P213 |
| Unit cell (Å) a, b, c | 145.1, 145.1, 145.1 |
| Unit cell (°) α, β, γ | 90, 90, 90 |
| No. of observations | 314,006 |
| No. of reflections | 25,520 (2,515) |
| Completeness (%) | 100.0 (100.0) |
| Mean I/σI | 39.2 (3.4) |
| CC1/2 | (0.890) |
| Rmergea | 0.066 (0.726) |
| Rpim | 0.020 (0.214) |
| Refinement statistics | |
| Resolution limits (Å) | 40.2–2.79 (2.91–2.79) |
| No. of reflections (work/free) | 25,461/1,292 |
| Completeness (%) | 98.9 (99.0) |
| Protein/ligand atoms | 7551/42 |
| Rcrystb | 0.206 (0.274) |
| Rfree (5.1% of data) | 0.240 (0.290) |
| Bonds (Å)/angles (°) | 0.003/0.508 |
| B-factors: protein/ligand (Å2) | 47.9/62.1 |
| Ramachandran plot statistics (%) | |
| Favored | 95.4 |
| Allowed | 4.3 |
| Outliers | 0.3 |
| MolProbity score | 1.95, 99th percentile (n = 4,464, 2.79 ± 0.25 Å) |
a Rmerge = ΣhklΣi|I(hkl)i − 〈I(hkl)〉|/ΣhklΣi 〈I(hkl)i〉.
b Rcryst = Σhkl|Fo(hkl) − Fc(hkl)|/Σhkl|Fo(hkl)| where Fo and Fc are observed and calculated structure factors, respectively.
Figure 1.
Crystal structure of domain-alternated S. pombe Uba1. A, comparison of the Uba1SCCH_ALT (left) and Uba1SCCH_OPEN (Protein Data Bank code 4II3, right) structures, shown as ribbon diagrams. Uba1 domains are labeled and color-coded, and ATP and Ub(a) from the Uba1SCCH_OPEN structure are shown as spheres and a yellow ribbon diagram, respectively. Catalytic cysteines positions (Cys593) are indicated with an arrow, and the SCCH domain alternation is highlighted by a double-headed arrow. B, ribbon diagrams of the Uba1SCCH_ALT (left) and Uba1SCCH_OPEN (right) structures overlaid with the 2Fo − Fc electron density (green) for the SCCH domain of the Uba1SCCH_ALT structure. For clarity, the SCCH domain of the Uba1SCCH_ALT structure is shown as loops. The Uba1 domains, ATP, and Ub(a) are colored and labeled as in A. alt, domain-alternated SCCH conformation.
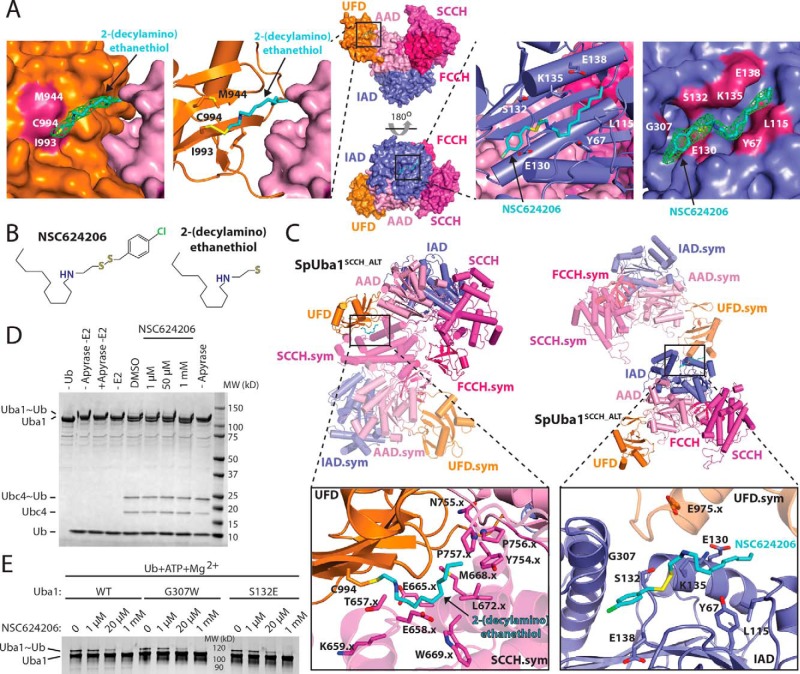
The final Uba1SCCH_ALT structure contains Uba1 residues 37–769 and 794–1012 and one ordered sulfate ion in a position that approximates where the α-phosphate of ATP normally binds Uba1. The only unaccounted for electron density in the maps was within a deep pocket on the IAD at the “bottom” of the Uba1 (Fig. 2A, right) and projecting off of the Cys994 side chain on the UFD (Fig. 2A, left). We were able to place the entire NSC624206 molecule into the electron density associated with the IAD (Fig. 2A, right). With regard to the site proximal to the UFD, because the electron density projects from the sulfhydryl group of Cys994, we surmised that the electron density belongs to the 2-(decylamino)ethanethiol fragment of NSC624206 (Fig. 2, A and B), which formed a disulfide-linked adduct to Cys994, with the (4-chlorophenyl)methanethiol fragment of NSC624206 serving as the leaving group.
Figure 2.
Interactions between the Uba1SCCH_ALT and NSC624206. A, middle, the Uba1SCCH_ALT structure is shown as a surface representation in two orientations, and the NSC624206 compound noncovalently bound to the IAD and the 2-(decylamino)ethanethiol fragment covalently bound to the UFD via disulfide bond are shown as sticks. The domains are colored and labeled as in Fig. 1. Left, magnified view of the 2-(decylamino)ethanethiol fragment of NSC624206, shown as sticks, bound to the UFD. Omit 2Fo − Fc electron density for the fragment is shown in green. The UFD and AAD are shown as surface representations, and residues that form contacts with the fragment are colored and labeled. Second from left, magnified view of the 2-(decylamino)ethanethiol fragment, shown as sticks, bound to Cys994 of the UFD, shown as a ribbon diagram. Side chains of residues that form contacts with the fragment are shown as sticks and labeled. The AAD is shown as a surface representation. Second from right, magnified view of the NSC624206 compound, shown as sticks, bound to the IAD, shown as a ribbon diagram. Side chains of residues that form contacts with the compound are shown as sticks and labeled. The IAD and SCCH are shown as surface representations. Right, magnified view of the NSC624206 compound, shown as sticks, bound to the IAD. Omit 2Fo − Fc electron density for the compound is shown in green. The IAD is shown as a surface representation, and residues that form contacts with the fragment are colored and labeled. B, chemical structures of the NSC624206 compound and the 2-(decylamino)ethanethiol fragment. C, crystal contacts with the NSC624206 compound. Top, the Uba1SCCH_ALT structure is shown as a ribbon diagram in two different orientations, and the domains are colored and labeled as in A. Symmetry (sym) mates that form contacts with the 2-(decylamino)ethanethiol fragment, shown as sticks, bound to the UFD (left) and to the NSC624206 compound, shown as sticks, bound to the IAD (right) are also shown as semitransparent ribbon diagrams with the domains colored and labeled. Bottom, magnified views of the 2-(decylamino)ethanethiol fragment bound to the UFD (left) and NSC624206 compound bound to the IAD (right). The side chains of residues from Uba1SCCH_ALT and the symmetry mates that form contacts with the fragment or compound are shown as sticks and labeled. The .x indicates residues residing in symmetry-related Uba1 molecules. D, biochemical analysis of the effect of 2-(decylamino)ethanethiol modification on UFD Cys994. An E1-E2 single turnover thioester transfer assay from Uba1 to Ubc4 was performed with different concentrations of NSC624206 or DMSO (see “Experimental procedures” for details). E, biochemical analysis of the inhibitory effect of NSC624206 on WT and mutant Uba1 activity. Uba1∼Ub thioester formation assays were performed as readouts of Uba1 catalytic activity with different concentrations of NSC624206 or DMSO (see “Experimental procedures” for details).
We next performed a series of biochemical experiments to determine whether the interactions between NSC624206 and Uba1 that we observed in our structure are related to the inhibitory mechanism (Fig. 2, D and E). First, to test whether binding of NSC624206 to the IAD as observed in our structure plays a role in its ability to inhibit Uba1, we analyzed the detailed intermolecular interactions (Fig. 2C) and generated Uba1 G307W and S132E Uba1 mutants designed to block NSC624206 from binding to the IAD and subjected these mutants to Uba1∼Ub thioester formation assays. The results of these experiments show that these mutations do not have an effect on the ability of NSC624206 to inhibit Uba1 activity (Fig. 2E). Because the UFD is involved in recruitment of E2 to Uba1 during the next step of the Ub conjugation cascade (i.e. transfer of Ub from Uba1 to the E2 catalytic cysteine), we wondered whether adduct formation between Cys994 of the UFD and the 2-(decylamino)ethanethiol fragment of NSC624206 might inhibit thioester transfer of Ub from E1 to E2 due to steric occlusion of the incoming E2. To test this, we used a single turnover experiment in which fully charged Uba1 was treated with apyrase (to prevent further Uba1∼Ub thioester formation) and NSC624206 followed by addition of E2 and monitoring formation of the E2∼Ub product. The results of this single turnover E1-E2 Ub thioester transfer experiment indicate that treatment of Uba1 with NSC624206 does not inhibit the E1-E2 Ub thioester transfer step (Fig. 2D).This could be because the Cys994–2-(decylamino)ethanethiol adduct does not form on the timescale of the assay, which is on the order of minutes compared with days with the crystallization experiments, or because Cys994–2-(decylamino)ethanethiol adduct formation does not inhibit E2 binding. Based on 1) the above biochemical data (Fig. 2, D and E), 2) the NSC624206-binding sites on Uba1 that are distant from the Uba1 active site (Fig. 2A), 3) the involvement of NSC624206 and the 2-(decylamino)ethanethiol fragment in crystal contacts (Fig. 2C), and 4) the proposed mechanism by which NSC624206 inhibits Uba1 activity (25), we conclude that the inhibitor-binding sites observed in our structure are not related to NSC624206's mechanism of inhibition but rather that NSC624206 served as a chemical additive that facilitated crystallization and X-ray diffraction.
Domain alternation and active site remodeling in S. pombe Uba1
To date, five crystal structures of Uba1 have been reported in which all nine copies of Uba1 are observed with the SCCH domain in the open conformation with the catalytic cysteine 34–37 Å away from the C terminus of Ub (14, 19, 20, 22). Although previous studies suggest that the structural basis for catalysis of thioester bond formation by Uba1 is conserved with SUMO E1 and involves domain alternation and active site remodeling (21), there is a lack of structural evidence supporting this hypothesis.
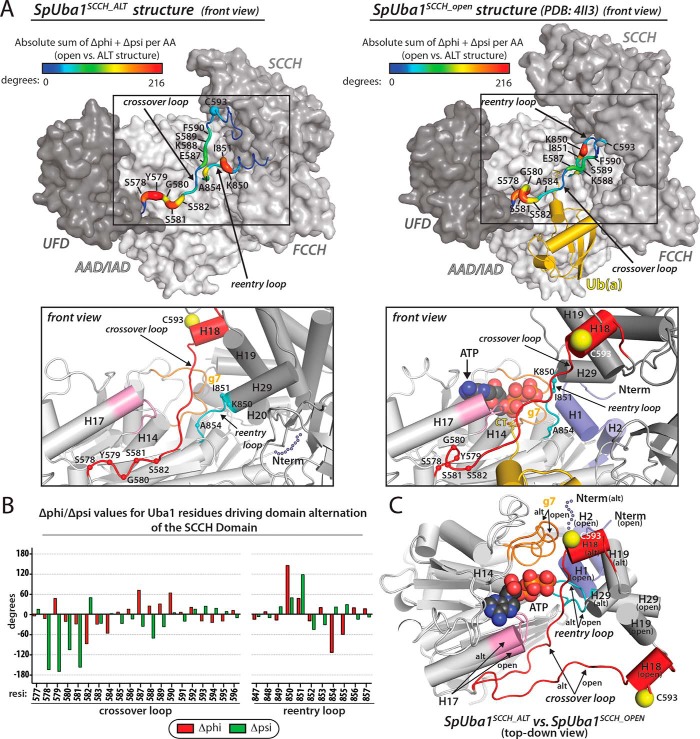
Comparison of the Uba1SCCH_ALT structure with previous Uba1 structures with the SCCH domain in the open conformation (hereafter referred to as Uba1SCCH_OPEN) (14, 19, 20, 22) reveals that the SCCH domain has undergone a domain alternation defined by a 106° rigid body rotation that translocates the catalytic cysteine residue (Cys593) 34 Å (Fig. 1A). Significant conformational changes accompanying SCCH domain alternation are also observed in the crossover and reentry loops that tether the SCCH domain to the AAD (Fig. 3, A–C) with the greatest structural changes in the crossover loop occurring at positions Ser578, Tyr579, Gly580, Ser581, and Ser582 and the greatest changes in the reentry loop occurring at positions Lys850, Ile851, and Ala854. Consistent with our structural observations, a previous study demonstrated that a K850P mutation in Uba1 significantly diminishes Uba1∼Ub thioester bond formation (21), presumably by hindering conformational flexibility required for SCCH domain alternation.
Figure 3.
Active site remodeling in S. pombe Uba1. A, conformational changes in the crossover and reentry loops upon SCCH domain alternation. Top, Uba1 from the Uba1SCCH_ALT and Uba1SCCH_open structures is shown as a surface representation with indicated domains colored various shades of gray. Ub(a) from the Uba1SCCH_OPEN structure is shown as a gold ribbon diagram. The crossover and reentry loops are shown as putty representations, and the diameter and color (see color code above structures) of the putty correspond to the sum of the absolute change in φ and ψ angles. Bottom, active site remodeling accompanies Uba1 SCCH domain alternation. The Uba1SCCH_ALT (left) and Uba1SCCH_OPEN (right) structures are shown in the same orientation as ribbon diagrams with domains colored as in the top panels. Regions undergoing conformational changes in the Uba1 active site upon SCCH domain alternation are color-coded and labeled. H1 and H2 of Uba1 that become disordered in the Uba1SCCH_ALT structure are shown as semitransparent slate spheres. The α-carbons of the residues that undergo significant changes between the open and closed conformations are labeled and shown as spheres. B, differences in φ and ψ angles for residues in the crossover and reentry loops between the Uba1SCCH_ALT and Uba1SCCH_OPEN structures were calculated and plotted. C, the adenylation domains of the Uba1SCCH_ALT and Uba1SCCH_OPEN structures were superimposed and are shown as ribbon diagrams. ATP from the Uba1SCCH_OPEN structure is shown as spheres. Note that this panel is presented in a top-down view, which differs from the front views presented in A to better highlight the conformational changes that occur during active site remodeling and SCCH domain alternation in the context of the superimposed structures. AA, amino acid; alt, domain-alternated SCCH conformation.
Also similar to the case for SUMO E1, comparison of the Uba1SCCH_ALT and Uba1SCCH_OPEN structures reveals remodeling of several elements of the adenylation active site (Fig. 3, A and C). The N-terminal helices of Uba1 (H1 and H2), which harbor residues crucial for adenylation, including Arg22 that coordinates the γ-phosphate of ATP (20–22), become disordered in the Uba1SCCH_ALT structure (Fig. 3, A and C). In addition to promoting disassembly of the adenylation active site, disordering of Uba1 H1 and H2 is necessary to accommodate domain alternation of the SCCH domain as major steric clashes would occur in the absence of conformational changes in these helices, in particular between H20, H28, and the reentry loop of the SCCH domain and H1 and H2 of the IAD. Additional elements of the adenylation active site observed to undergo remodeling in the Uba1SCCH_ALT structure include the 310 helix g7 (Fig. 3, A and C), which harbors residues Asn471, Leu472, and Arg474 known to play a key role in adenylation (21). In the Uba1SCCH_ALT structure, g7 projects away from the ATP·Mg binding site, positioning residues involved in catalysis such that they are incapable of interacting with ATP·Mg. Furthermore, a change in the relative angle of helix H17 of the AAD (Fig. 3C) positions residues Leu536, Asn538, and Ala541 such that they are incompatible with ATP binding due to major steric clashes that would occur. Asp537, which is involved in Mg2+ coordination and is essential for Uba1 activity, is also positioned suboptimally in the Uba1SCCH_ALT structure.
Importantly, the changes observed in the adenylation active site are unique to the Uba1SCCH_ALT structure as each of these elements adopt similar conformations in all Uba1SCCH_OPEN structures. Taken together with the fact that the Uba1SCCH_ALT structure was determined in the absence of Ub/ATP·Mg and does not harbor a suicide inhibitor designed to trap the tetrahedral intermediate generated during E1∼Ub thioester bond formation such as 5′-(vinylsulfonylaminodeoxy)adenosine (21), our observations suggest that domain alternation and remodeling of the adenylation active site are interconnected and intrinsic structural features of Uba1 that do not require substrate binding and catalysis to occur. Thus, it is likely that the open and closed conformations of Uba1 exist in equilibrium in the absence of ATP·Mg and Ub. Our structural observations are consistent with a model in which ATP binding to Uba1 promotes assembly of the adenylation active site, including ordering of helices H1 and H2, which shifts the equilibrium to the open state due to clashes between the SCCH domain and H1/H2 that would occur when the adenylation active site is assembled. As noted previously, the release of pyrophosphate after adenylation of the Ub C terminus (the rate-determining step of the reaction (4)) promotes disorder of H1/H2 helices, which shifts the open/closed SCCH equilibrium in the closed direction, which is required for thioester bond formation (21).
Comparison of S. pombe Uba1 and SUMO E1 structures
Comparison of SUMO E1 and Uba1 structures reveals similar overall open conformations in which the catalytic cysteine is separated from the Ubl C terminus by 34–37 Å. Although the extent of the SCCH domain alternation differs slightly between the SUMO E1SCCH_CLOSED and Uba1SCCH_ALT structures (130° and 106°, respectively), the general features of the rigid body rotation are similar, including the direction of the rotation and the regions in the crossover and reentry loops that accommodate this conformational change (Fig. 4, A–C). Notably, reentry loop residues that undergo the greatest conformational changes during SCCH domain alternation in SUMO E1 (Gly381 and Asn382) correspond to those of Uba1 (Lys850 and Ile851). In contrast, Uba1 crossover loop residues that undergo the greatest conformational changes during SCCH domain alternation are offset by five residues compared with those in SUMO E1 where the greatest changes are in residues Lys164–Arg168 (21). This difference is likely due to the fact that Cys159 and Cys161 of SUMO E1, which correspond to Ser578 and Ser581 of Uba1, are involved in zinc coordination, a feature unique to SUMO and NEDD8 E1s that reduces conformational flexibility in this region of the crossover loop (15, 17). Although the path of the crossover loop in the Uba1SCCH_ALT structure places the Uba1 catalytic cysteine ∼23 Å from the where the C terminus of Ub would be located (Fig. 4, A and C), in the context of the observed SCCH domain alternation, Cys593 can be positioned proximal to the electrophilic center simply through conformational changes within the highly flexible crossover loop and melting of the α helix harboring Cys593 (H18) as in SUMO E1.
Figure 4.
Comparison of Uba1 and SUMO E1 in different conformational states. A, the Uba1SCCH_OPEN (left) and Uba1SCCH_ALT (right) structures are shown as surface representations (with the exception of the SCCH domain, which is shown as a light magenta ribbon diagram) with domains color-coded and labeled. Ub(a) and ATP were modeled onto the Uba1SCCH_ALT structure based on how they interact with Uba1 in the Uba1SCCH_OPEN structure. SCCH domain helices are labeled, and the N- and C-terminal ends of the helices are indicated to facilitate structure comparison. H18 and H19 of the Uba1 structures are colored red to correspond with H6 and H7 of the SUMO E1 structures in B. H29 of the Uba1 structures is colored cyan to correspond with H13 of the SUMO E1 structures in B. The approximate direction of the SCCH domain alternation is indicated with a double-headed arrow. A magnified view of the Uba1SCCH_ALT active site is shown in the right inset. B, the SUMO E1SCCH_OPEN (Protein Data Bank code 3KYC) and SUMO E1SCCH_CLOSED (Protein Data Bank code 3KYD) structures are presented in the same style and orientation as Uba1 in A. C, comparison of the crossover/reentry loops and SCCH domains in the Uba1SCCH_OPEN (Protein Data Bank code 4II3) and SUMO E1SCCH_OPEN (Protein Data Bank code 3KYC) structures (bottom) and in the Uba1SCCH_ALT and SUMO E1SCCH_CLOSED (Protein Data Bank code 3KYD) structures (top). The adenylation domains of the structures were superimposed. The structures are shown as ribbon diagrams and ATP, 5′-(vinylsulfonylaminodeoxy)adenosine (AVSN), or 5′-(sulfamoylaminodeoxy)adenosine (AMSN) are shown as sticks. The difference in SCCH domain positioning in the Uba1SCCH_ALT and SUMO E1SCCH_CLOSED structures is indicated with a double-headed arrow. D, left, comparison of the Uba1 and SUMO E1 SCCH domains. Unique structural elements of the Uba1 SCCH domain are indicated with a black dashed oval. Right, the Uba1 SCCH domain was docked onto Uba1 in the same orientation as the SUMO E1SCCH_CLOSED structure. Steric clashes involving the unique structural elements of the Uba1 SCCH domain illustrate the fact that the extent of domain alternation in Uba1 and SUMO E1 during thioester bond formation is unlikely to be precisely the same. alt, domain-alternated SCCH conformation; SUMO(a), adenylated SUMO.
Although the SCCH domains of Uba1 and SUMO E1 harbor a conserved globular core, there are several unique structural features of the Uba1 SCCH domain that are worth noting in the context of the slight difference in the extent of SCCH domain rotation observed in the Uba1SCCH_ALT and SUMO E1SCCH_CLOSED structures. First, the H24-H25 loop of Uba1 harbors an 11-residue insertion compared with the corresponding loop of SUMO E1 (the H9-H10 loop) (Fig. 4D). Furthermore, H21 and H22 are unique to Uba1 as the corresponding region of SUMO E1 SCCH lacks helicity and is disordered in all available structures (Fig. 4D). Modeling the Uba1 SCCH domain in the precise conformation observed in the SUMO E1SCCH_CLOSED structure reveals that the unique H24-H25 loop and H21/H22 insertions of Uba1 would severely clash with the AAD and IAD, respectively (Fig. 4D). This suggests that the magnitude of SCCH domain alternation required for thioester bond formation in Uba1 and SUMO E1 is unlikely to be precisely the same. With that said, the SCCH domain extensively covers the SUMO E1 active site in the closed conformation, including contacts between the H9-H10 loop and the AAD (Fig. 4B), whereas in the Uba1SCCH_ALT structure, the SCCH domain less extensively covers the active site, and the H24-H25 loop does not engage in contacts to the AAD (Fig. 4A). Whether this reflects actual differences in the bona fide Uba1SCCH_CLOSED structure or indicates the need for a slight additional rotation of the SCCH domain compared with what we observed in the Uba1SCCH_ALT structure awaits structural characterization of the tetrahedral intermediate formed during E1∼Ub thioester bond formation. This slight additional rotation of the SCCH domain (if necessary) could easily be achieved through the additional conformational changes in the flexible crossover and reentry loops necessary to properly position the catalytic cysteine for nucleophilic attack during thioester bond formation.
Domain alternation results in distinct networks of intramolecular contacts between the SCCH domain and other Uba1 domains
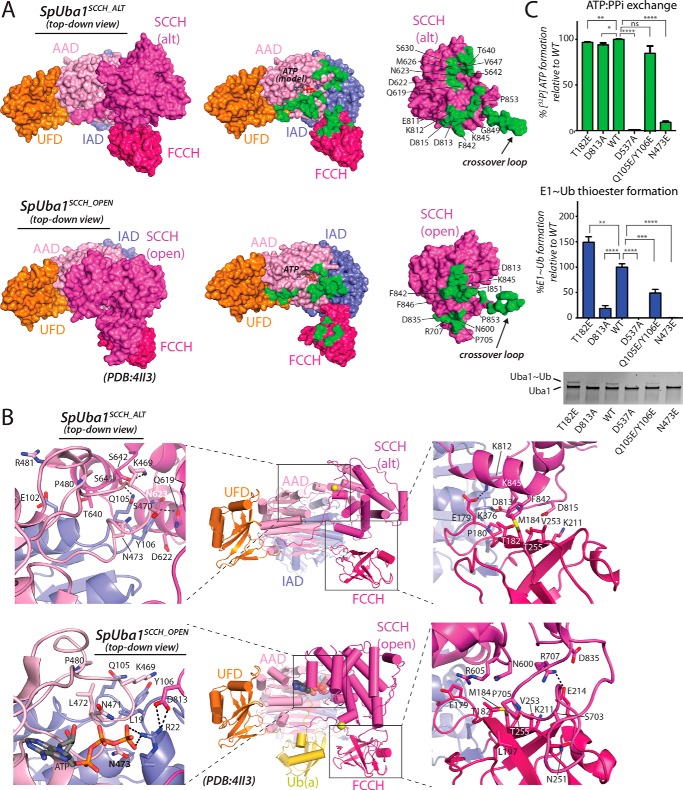
Although a total of ∼2300 Å2 of surface area is buried at the intramolecular interface between the SCCH domain and the AAD, IAD, and FCCH domain in Uba1SCCH_OPEN structures, SCCH domain alternation and the resulting network of unique intramolecular interactions increases this value to ∼3200 Å2 in the Uba1SCCH_ALT structure (Fig. 5, A and B). Interestingly, although there are relatively few contacts between the FCCH and SCCH domains in Uba1SCCH_OPEN structures, the buried surface at the interface between these two domains nearly doubles (∼700 Å2) in Uba1SCCH_ALT (Fig. 5A). Specifically, during SCCH domain alternation, a highly conserved salt bridge between Glu214 of the FCCH domain and Arg707 of the SCCH domain observed in Uba1OPEN structures is broken, and a completely different network of contacts forms, including a salt bridge between Glu179 of the FCCH domain and Lys812 of the SCCH domain (Fig. 5B, right panels). Additionally, Phe842 of the SCCH domain inserts into a hydrophobic patch on the FCCH domain formed by Thr182, Met184, and Val235. This suggests that rather than being conformationally flexible upon SCCH domain alternation the FCCH domain of Uba1 may contribute to thioester bond formation by stabilizing the SCCH closed conformation via the formation of these additional intramolecular contacts. Although Thr182 does not form any intramolecular contacts in the Uba1SCCH_OPEN structure, it does form the aforementioned contact with Phe842 in the Uba1SCCH_ALT structure. A T182E mutation results in a very slight decrease in Ub adenylate formation but modestly increases E1∼Ub thioester formation (Fig. 5C). The larger, charged glutamic acid side chain can presumably form more contacts with residues in the SCCH, which would further support the idea that the FCCH domain stabilizes the SCCH closed conformation during thioester transfer. Notably, this appears to be a feature unique to Uba1 as the FCCH domain of SUMO E1 is significantly smaller and largely disordered in all SUMO E1 structures, including SUMO E1SCCH_CLOSED (15, 21) (Fig. 4B).
Figure 5.
A unique network of contacts among the Uba1 SCCH, FCCH, and AAD domains in the Uba1SCCH_ALT structure. A, Uba1SCCH_ALT (top left) and Uba1SCCH_OPEN (bottom left) structures are shown as surface representations with domains labeled and color-coded. For clarity, Ub(a) from the Uba1SCCH_OPEN structure is not shown. The right panels show the intramolecular interface between Uba1 and the globular SCCH domain as an open book representation with residues buried at the interface shaded green. To highlight the altered network of interactions resulting from the SCCH domain alternation, SCCH residues involved in unique intramolecular interactions in the two structures are labeled. B, Uba1SCCH_ALT (top) and Uba1SCCH_OPEN (bottom) structures are shown as ribbon diagrams with magnified views of the SCCH/AAD and SCCH/FCCH intramolecular interfaces shown in the left and right insets, respectively. Residues involved in interactions are shown as sticks, and hydrogen bonds are indicated with dashed black lines. C, structure-function analysis of intramolecular contacts between Uba1 and the globular SCCH domain unique to the Uba1SCCH_ALT structure. ATP/PPi exchange assays (top) and Uba1∼Ub thioester formation assays (bottom) were performed as readouts of Uba1 catalytic activities. All experiments were performed in triplicate as described under “Experimental procedures,” and error bars represent ±1 S.D. *, p ≤ 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant. alt, domain-alternated SCCH conformation.
Domain alternation also results in a completely different network of intramolecular interactions between the SCCH domain and IAD/AAD in the Uba1SCCH_ALT structure with most changes occurring around the Uba1 active site. A total of ∼850 Å2 of surface area is buried at the interface between the SCCH and AAD/IAD in Uba1SCCH_OPEN structures, most of which (∼650 Å2) involves H1 and H2 of the IAD that become disordered in the Uba1SCCH_ALT structure (Fig. 5, A and B). The H1 and H2 active site remodeling that occurs during SCCH domain alternation significantly affects interdomain contacts in this region. Asn473 of the AAD, located on helix g7, forms a hydrogen bond with Gln23 of helix H1 in the Uba1SCCH_OPEN structure but also forms contacts with Gln105 and Tyr106 in the Uba1SCCH_ALT structure (Fig. 5B, left panels). As expected, an N473E mutation decreases both adenylation and E1∼Ub thioester formation activity compared with wild type (WT) (Fig. 5C), confirming this residue's role in domain stabilization in both the open and closed conformations.
Other notable interactions resulting from domain alternation involve Ser641 and Ser642 of the SCCH domain that engage in hydrogen bonds to Gln105 of the IAD and Lys469 of the AAD, respectively. Gln619, Asp622, and Asn623 of the SCCH are also brought in proximity to Tyr106 of the AAD where a network of van der Waals interactions takes place (Fig. 5B, left panels). Consistent with these observations, a Q105E/Y106E double mutant, which disrupts a major set of interactions in the Uba1SCCH_ALT structure but should not have any consequence based on the Uba1SCCH_OPEN structure, has no effect on Ub adenylation, but it does show a 2-fold decrease in Uba1∼Ub thioester formation (Fig. 5C), suggesting a defect specifically at the thioester bond formation step of Ub activation. Additionally, in the Uba1SCCH_OPEN structure, Asp813 from the SCCH domain forms a salt bridge with Arg22 from the IAD, which contacts the γ-phosphate of ATP; however, in the Uba1SCCH_ALT structure, Asp813 contacts Lys376 from the IAD domain and Pro180 from the FCCH domain (Fig. 5B, right panels). A D813A mutation does not have an effect on adenylation activity but does result in a loss of E1∼Ub thioester formation (Fig. 5C). Thus, the contacts between Asp813 and specific residues in the IAD and FCCH domains could be important for stabilizing the E1 during SCCH alternation during thioester bond formation. Finally, a D537A Uba1 mutant that is known to lack catalytic activity due to an inability to coordinate a magnesium ion necessary for adenylation was used as a negative control for the adenylation and thioester formation assays (Fig. 5C).
Conclusions
Here, we have presented the first crystallographic snapshot of Uba1 in which SCCH domain alternation and active site remodeling have been observed. Although precise details await determination of a bona fide Uba1∼Ub–AMP tetrahedral intermediate structure, our Uba1SCCH_ALT structure provides the first structural evidence that the salient features of thioester bond formation by Uba1 are shared with SUMO E1. Our data suggest that SCCH domain alternation and active site remodeling are interconnected and intrinsic structural features of Uba1 and that the open and closed conformations of Uba1 exist in equilibrium prior to ATP·Mg and Ub binding. Our structural observations, together with previous data, are consistent with a model in which ATP binding to Uba1 shifts the equilibrium to an open state (adenylation active) and that pyrophosphate release after adenylation shifts the equilibrium to the closed state (thioester active). Finally, the insights provided by the novel conformational snapshot of Uba1 presented in this study may guide efforts to develop small molecule inhibitors of this critically important enzyme that is an active target for anticancer therapeutics.
Experimental procedures
Protein expression, purification, and crystallization
WT and mutated variants of full-length (residues 1–1012) S. pombe Uba1, S. pombe Ubc4, and S. pombe Ub (residues 1–76) used in structural and biochemical studies were expressed in Escherichia coli and purified as described previously (19). Purified proteins were concentrated to ∼25 (Uba1 WT and variants) or ∼10 mg/ml (Ub) and snap frozen in liquid nitrogen for use in subsequent crystallization and biochemical experiments. The sample used for crystallization and subsequent determination of the Uba1SCCH_ALT structure was obtained by mixing Uba1C593A mutant protein (177 μm final), the Uba1 inhibitor NSC624206 (1 mm final from a 50 mm stock in 100% DMSO; Tocris Bioscience), and MgCl2 (5 mm final). Following a 30-min incubation on ice, 1 μl of the Uba1C593A/NSC624206/MgCl2 mixture (in 20 mm Tris, pH 8.0, ∼250 mm NaCl buffer) was added to 1 μl of crystallization buffer (80 mm sodium citrate, pH 5.9, 1 m ammonium sulfate) followed by hanging drop vapor diffusion at 15 °C. Reducing agent was omitted to maintain the integrity of NSC624206, which contains a disulfide bond. Uba1SCCH_ALT crystals grew to maximum size in 2 weeks and were flash frozen in liquid nitrogen following cryoprotection in an 80 mm sodium citrate, pH 5.9, 4 m ammonium sulfate solution.
X-ray data collection, structure determination, and refinement
A complete data set for the Uba1SCCH_ALT crystals was collected to a resolution of 2.79 Å at the Advanced Photon Source (Argonne, IL), Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID. All data were indexed, integrated, and scaled using HKL2000 (26). The Uba1SCCH_ALT crystals belong to space group P213 with unit cell dimensions a = b = c = 145.1 Å, and there is one molecule of Uba1 per asymmetric unit. Initial molecular replacement efforts using complete existing Uba1 structures as search models failed. A search model comprising Uba1 from the S. pombe Uba1/Ub/ATP·Mg complex (Protein Data Bank code 4II3) (20) with the SCCH domain deleted yielded a successful molecular replacement solution using the program Phaser (27). After one round of refinement, the resulting maps were inspected, and clear electron density for the SCCH domain in a conformation significantly different from that in previous structures was evident. A subsequent multiple ensemble molecular replacement search comprising 1) the previous molecular replacement solution and 2) the standalone SCCH domain resulted in successful placement of the SCCH domain into the appropriate electron density. The model was refined to R/Rfree values of 0.206/0.240 via iterative rounds of refinement and rebuilding using PHENIX (28) and Coot (29). All molecular graphics representations of the structures were generated using PyMOL (30).
The final Uba1SCCH_ALT structure contains Uba1 residues 37–769 and 794–1012, and there is one ordered sulfate ion in a position that approximates where the α-phosphate of ATP normally binds Uba1. As detailed under “Structure determination and characterization of a Schizosaccharomyces pombe Uba1/NSC624206 co-complex”, electron density for an intact NSC624206 inhibitor noncovalently bound to the Uba1 IAD as well as a 2-(decylamino)ethanethiol fragment of NSC624206 that formed a disulfide-linked adduct to Cys994 of UFD was evident (also see Fig. 2, A–C).
ATP/PPi isotope exchange assays
Radioactive isotope-based ATP/PPi exchange assays were adapted from previous work (31, 32). 50-μl reaction mixtures containing 50 mm Tris, pH 7.5, 10 mm MgCl2, 0.5 mm DTT, 1 mm ATP, 1 mm [32P]PPi (PerkinElmer Life Sciences), and 10 μm Ub were incubated with 0.2 μm WT or mutant E1 proteins at 37 °C for 20 min before the reaction was quenched with 5% (w/v) trichloroacetic acid (0.5 ml) containing 4 mm carrier PPi. After processing, data were quantitated by Cerenkov counting.
Uba1∼Ub thioester formation assays
Uba1∼Ub thioester formation assays for wild-type and mutant proteins were performed in a reaction containing 50 nm Uba1, 2 μm Ub, 5 mm MgCl2, 20 mm MES, pH 5.5, 50 mm NaCl, and 2 μm ATP. Reactions were incubated for 5 s at 4 °C. For inhibitor-mediated thioester transfer assays, 1 μm, 20 μm, or 1 mm NSC624206 inhibitor was added to the reactions, which were then incubated for 30 s at room temperature. To account for DMSO required to solubilize NSC624206, 5% DMSO was included in all assays involving inhibitor. After incubation, all reactions were denatured in non-reducing SDS-PAGE buffer and subjected to SDS-PAGE. The gels were stained with SYPRO Ruby (Bio-Rad) and visualized with a ChemiDoc MP (Bio-Rad).
Author contributions
Z. L., G. A.-M., and S. K. O. performed structural and biochemical experiments. Z. L., L. Y., J. H. A., G. A.-M., Y. C., and S. K. O. analyzed data. Z. L., J. H. A., Y. C., and S. K. O. prepared the manuscript.
Acknowledgments
We thank Christopher Davies and Katelyn Williams for critically reading the manuscript. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by National Institute of General Medical Sciences, National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on the 24-ID-C beamline is funded by National Institutes of Health Office of Research Infrastructure Program High-End Instrumentation Grant S10 RR029205). This research used resources of the Advanced Photon Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. This work is also based upon research conducted at Southeast Regional Collaborative Access Team beamline 22-ID. The X-ray crystallography facility used for this work is supported by Office of the Vice President for Research at the Medical University of South Carolina. The crystallization robotics instrument used in this study was purchased via National Institutes of Health Shared Instrumentation Award S10 RR027139-01.
This work was supported by National Institutes of Health Grants R01 GM115568 (to S. K. O.) and R01 CA088932 (to Y. C.) and in part by Hollings Cancer Center T32 Ruth L. Kirschstein National Research Service Award Training Program Grant T32 CA193201 (to J. A.) from the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 5UM6) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Ub
- ubiquitin
- Ubl
- ubiquitin-like modifier
- AAD
- active adenylation domain
- IAD
- inactive adenylation domain
- UFD
- ubiquitin fold domain
- FCCH domain
- first catalytic cysteine half-domain
- SCCH domain
- second catalytic cysteine half-domain
- SUMO
- small ubiquitin-like modifier
- Uba1SCCH_ALT
- Uba1 “closed” conformation
- Uba1SCCH_OPEN
- Uba1 open conformation (Protein Data Bank code 4II3)
- SUMO E1SCCH_CLOSED
- SUMO E1 closed conformation (Protein Data Bank code 3KYD)
- SUMO E1SCCH_OPEN
- SUMO E1 open conformation (Protein Data Bank code 3KYC)
- Ub(a)
- ubiquitin adenylate (i.e. ubiquitin adenylated at its C-terminal glycine residue).
References
- 1. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 2. van der Veen A. G., and Ploegh H. L. (2012) Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 [DOI] [PubMed] [Google Scholar]
- 3. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haas A. L., and Rose I. A. (1982) The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 5. Haas A. L., Warms J. V., Hershko A., and Rose I. A. (1982) Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 257, 2543–2548 [PubMed] [Google Scholar]
- 6. Haas A. L., Bright P. M., and Jackson V. E. (1988) Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J. Biol. Chem. 263, 13268–13275 [PubMed] [Google Scholar]
- 7. Hershko A., Heller H., Elias S., and Ciechanover A. (1983) Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 8. Pickart C. M., and Rose I. A. (1985) Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260, 1573–1581 [PubMed] [Google Scholar]
- 9. Hänzelmann P., Schäfer A., Völler D., and Schindelin H. (2012) Structural insights into functional modes of proteins involved in ubiquitin family pathways. Methods Mol. Biol. 832, 547–576 [DOI] [PubMed] [Google Scholar]
- 10. Berndsen C. E., and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 [DOI] [PubMed] [Google Scholar]
- 11. Schulman B. A., and Harper J. W. (2009) Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stewart M. D., Ritterhoff T., Klevit R. E., and Brzovic P. S. (2016) E2 enzymes: more than just middle men. Cell Res. 26, 423–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Streich F. C. Jr., and Lima C. D. (2014) Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee I., and Schindelin H. (2008) Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 [DOI] [PubMed] [Google Scholar]
- 15. Lois L. M., and Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L. Jr., Holton J. M., and Schulman B. A. (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 17. Walden H., Podgorski M. S., and Schulman B. A. (2003) Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422, 330–334 [DOI] [PubMed] [Google Scholar]
- 18. Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., and Schulman B. A. (2007) Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lv Z., Rickman K. A., Yuan L., Williams K., Selvam S. P., Woosley A. N., Howe P. H., Ogretmen B., Smogorzewska A., and Olsen S. K. (2017) S. pombe Uba1-Ubc15 structure reveals a novel regulatory mechanism of ubiquitin E2 activity. Mol. Cell 65, 699.e6–714.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olsen S. K., and Lima C. D. (2013) Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol. Cell 49, 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olsen S. K., Capili A. D., Lu X., Tan D. S., and Lima C. D. (2010) Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schäfer A., Kuhn M., and Schindelin H. (2014) Structure of the ubiquitin-activating enzyme loaded with two ubiquitin molecules. Acta Crystallogr. D Biol. Crystallogr. 70, 1311–1320 [DOI] [PubMed] [Google Scholar]
- 23. Brownell J. E., Sintchak M. D., Gavin J. M., Liao H., Bruzzese F. J., Bump N. J., Soucy T. A., Milhollen M. A., Yang X., Burkhardt A. L., Ma J., Loke H. K., Lingaraj T., Wu D., Hamman K. B., et al. (2010) Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell 37, 102–111 [DOI] [PubMed] [Google Scholar]
- 24. Chen J. J., Tsu C. A., Gavin J. M., Milhollen M. A., Bruzzese F. J., Mallender W. D., Sintchak M. D., Bump N. J., Yang X., Ma J., Loke H. K., Xu Q., Li P., Bence N. F., Brownell J. E., et al. (2011) Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. J. Biol. Chem. 286, 40867–40877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ungermannova D., Parker S. J., Nasveschuk C. G., Chapnick D. A., Phillips A. J., Kuchta R. D., and Liu X. (2012) Identification and mechanistic studies of a novel ubiquitin E1 inhibitor. J. Biomol. Screen 17, 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Delano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 31. Burch T. J., and Haas A. L. (1994) Site-directed mutagenesis of ubiquitin. Differential roles for arginine in the interaction with ubiquitin-activating enzyme. Biochemistry 33, 7300–7308 [DOI] [PubMed] [Google Scholar]
- 32. Wang J., Cai S., and Chen Y. (2010) Mechanism of E1-E2 interaction for the inhibition of Ubl adenylation. J. Biol. Chem. 285, 33457–33462 [DOI] [PMC free article] [PubMed] [Google Scholar]