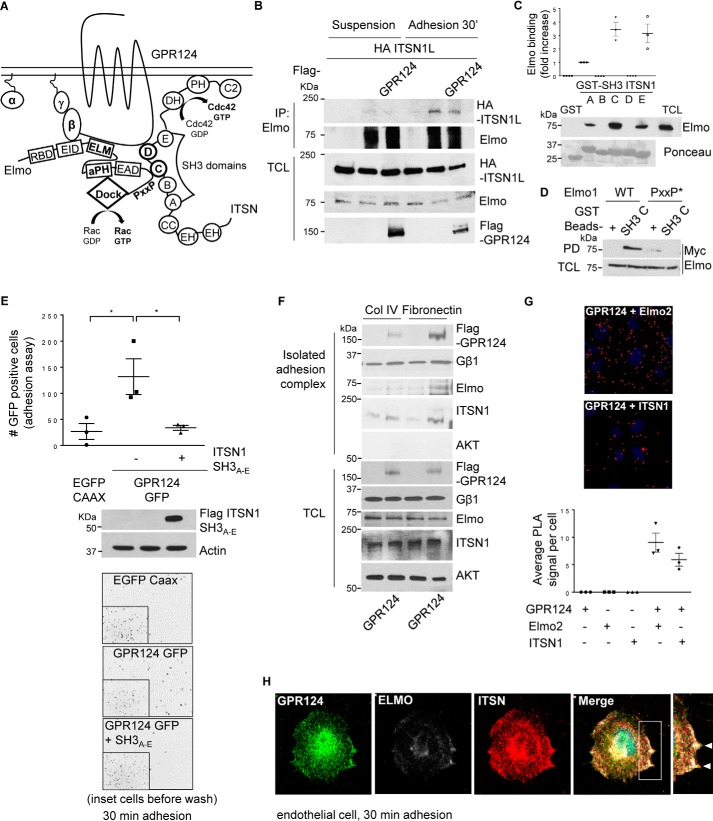
Figure 5.
GPR124-dependent cell adhesion is mediated by its interaction with ITSN, which directly interacts with Elmo–Dock180, forming a signaling complex that co-localizes with GPR124. A, working model. GPR124 interacts with the Gβγ–Elmo–Dock complex as well as with ITSN1/2. These atypical and conventional guanine nucleotide exchange factors also directly interact with each other, constituting a novel signaling complex that mediates GPR124-dependent cell adhesion. B, Elmo interacts with ITSN during cell adhesion. COS7 cells expressing full-length HA–ITSN1 and FLAG–GPR124 (as indicated) were left in suspension or adhering for 30 min. Then endogenous Elmo was immunoprecipitated, and interacting HA-ITSN1 was revealed by Western blotting. Preimmune rabbit IgG was used as a negative control for immunoprecipitation (first lane for suspension and adhesion conditions). C, endogenous Elmo directly interacts with ITSN1-SH3A, ITSN1-SH3C, and ITSN1-SH3E domains. HEK293T cell lysates were incubated for 3 h at 4 °C with individual recombinant GST–ITSN1-SH3 domains (A–E). GST pulldown assays were performed as described under “Experimental procedures.” Endogenous Elmo specifically binds to SH3A, SH3C, and SH3E domains of ITSN1 (PD, Western blotting, anti-Elmo). Recombinant GST–ITSN-SH3 domains are shown stained with Ponceau. D, the proline-rich (PXXP) motif in Elmo is required to bind the ITSN1-SH3C domain. Cell lysates from HEK293T cells expressing either wild-type Myc–Elmo or PXXP* mutant Myc–Elmo were incubated for 3 h at 4 °C with the recombinant GST–ITSN1-SH3C domain, and the interacting Elmo was revealed by Western blotting with anti-Myc (top). The expression of Elmo in total cell lysates (TCL) is shown at the bottom. GST was used as a negative control in the pulldown assays. The interaction between Elmo and ITSN1-SH3C domain was lost when the proline-rich region of Elmo was mutated. E, GPR124-dependent cell adhesion is inhibited by the ITSN1-SH3A–E module. COS7 cells were transfected with GPR124–GFP either with or without the FLAG–ITSN1-SH3A–E module. Cell adhesion assays were performed for 30 min. GPR124-dependent cell adhesion was decreased in FLAG–ITSN1-SH3A–E module-expressing cells. Basal adhesion of EGFP–CAAX–expressing cells was used as a reference. Bars, mean ± S.E. (error bars). Statistics were performed by one-way ANOVA followed by Tukey's multiple-comparison post hoc test (*, p < 0.05; n = 3). The middle panel shows the expression of FLAG–ITSN1-SH3A–E module in total cell lysates, and actin was used as a loading control. Representative images showing adherent cells are shown at the bottom; insets show all fluorescent cells in the field before washing out non-adherent cells. F, GPR124, Gβγ, Elmo, and ITSN are detected in the isolated adhesion complex. Control and GPR124-transfected COS7 cells were left to adhere for 30 min on collagen IV or fibronectin-coated plates. Adherent cells were lysed, and proteins that remained attached to the plates were washed and recovered with Laemmli sample buffer. GPR124, Gβγ, Elmo, and ITSN were detected by Western blotting in the isolated adhesion complex as well as in total cell lysates. AKT was used to confirm that isolated adhesion complexes were not contaminated by nonspecifically bound cytosolic proteins and as a loading control in TCL. G, endogenous interaction between GPR124 and Elmo2 as well as between GPR124 and intersectin 1 in endothelial cells. Proximity ligation assays were performed in endothelial cells (HUVECs) using the indicated pairs of antibodies to detect endogenous GPR124 interacting with endogenous Elmo or with endogenous ITSN1. Individual antibodies were used alone as a control. The PLA signal per cell was quantified by ImageJ software. At least 30 cells were analyzed. The graph represents three independent experiments (mean ± S.E., n = 3). Representative pictures of PLA signals, depicted as red dots, are shown in the top and middle panels. Cell nuclei were stained with DAPI. H, endogenous GPR124 co-localizes with Elmo and ITSN at cell protrusions of adhering endothelial cells. Cell adhesion assays were performed on gelatin-coated glass coverslips for 30 min, followed by immunostaining to determine the localization of endogenous GPR124, Elmo, and ITSN proteins in HUVECs (white arrows). A representative cell observed by confocal microscopy is shown. Similar results were observed in 29 of 61 cells.