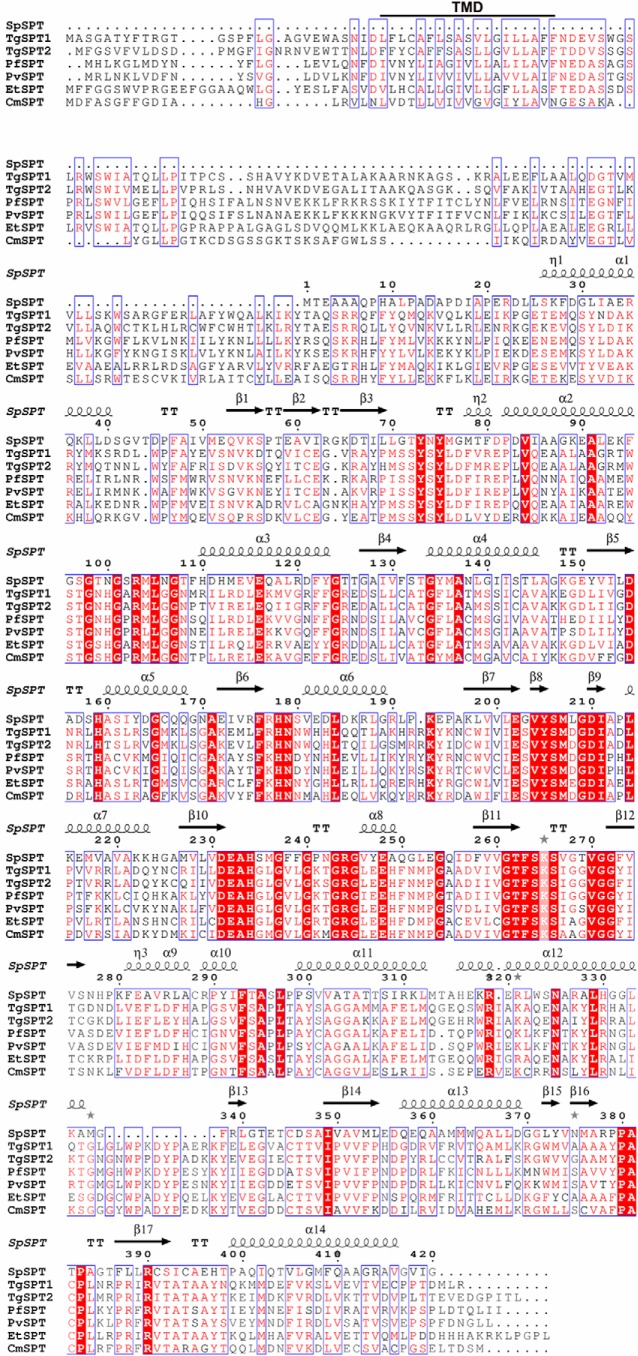
Figure 2.
Sequence alignment of the predicted SPT from four members of the Apicomplexa (T. gondii, TgSPT1 and TgSPT2; E. tenella, EtSPT; P. falciparum, PfSPT; and Plasmodium vivax, PvSPT) and the characterized enzyme from the prokaryote S. paucimobilis (SpSPT). Conserved residues (including those in the active site) identified by analyses of the SpSPT structure and homology modeling of the human functional orthologue (LCB2), are highlighted in red, with red text denoting similarity. Blue boxes denote conserved domains. The canonical lysine demonstrated to form an internal aldimine with the co-factor PLP at SpSPT position 265 is highlighted (21). The N-terminal extensions unique to the predicted apicomplexan enzymes harbor a transmembrane domain predicted by TMPRED (TMD, bold and underlined). The figure was produced using ESPript 3.0 (59).