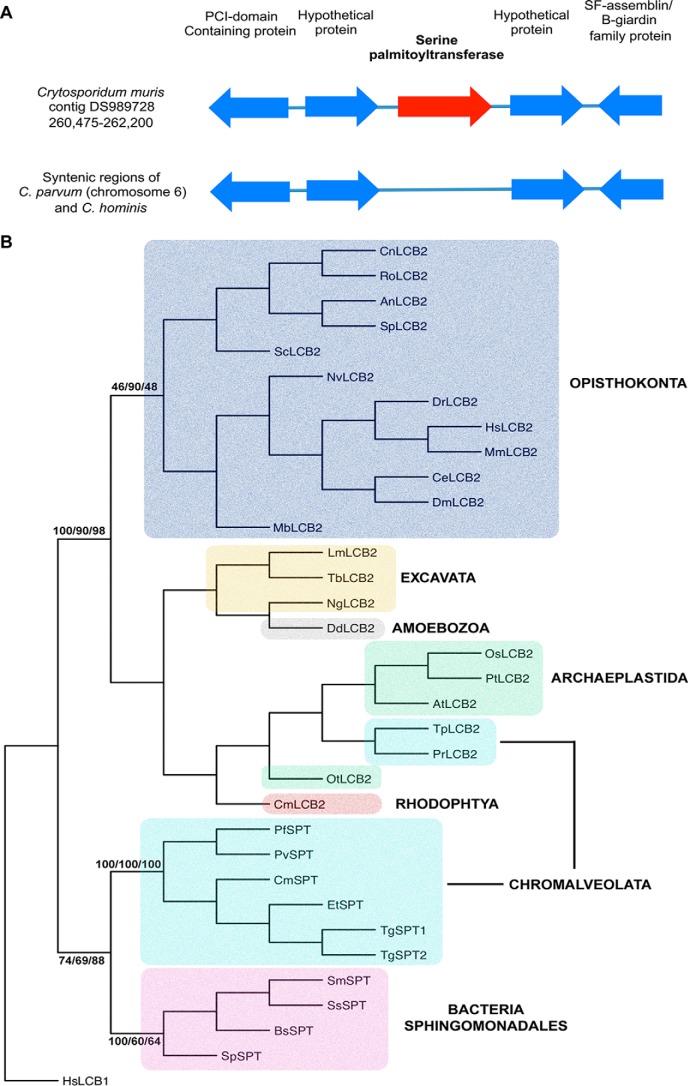
Figure 8.
A, schematic illustrating the gene arrangement in the region surrounding the encoded C. muris serine palmitoyltransferase, compared with the syntenic regions of C. parvum and C. hominis chromosome 6. B, phylogenetic tree produced from a genetic distance matrix showing the relationship between the eukaryotic catalytic subunit of serine palmitoyltransferase (LCB2) and the prokaryotic and apicomplexan orthologues (SPT). The Opistokonta (animals and fungi) are colored blue; the Excavata (subgroup of unicellular eukaryotes) are yellow; Amoebozoa (amoeboid protozoa) are gray; Archaeplastida (plants and algae, containing cyanobacterium-derived plastid) are green; Rodophyta (a subgroup of the Archaeplastida, red algae) are red; Chromalveolata (unicellular eukaryotes containing red algal derived plastid) are turquoise; and Sphingomonadales (alphaproteobacteria with the ability to synthesize sphingolipids) are pink. The bootstrap values of the major clades are shown where they are greater than 60 at common nodes for the three methodologies employed: F-MDist, RAxML, and PhyML. The non-catalytic subunit of the human serine palmitoyltransferase (HsLCB1) was utilized as the outgroup. Sequences used in the analyses were equivalent to those aligned to TgSPT1 amino acids 228–411. Sequence information: LCB1, serine palmitoyltransferase subunit 1; LCB2, palmitoyltransferase subunit 2; SPT, serine palmitoyltransferase. NBCI accession numbers: HsLCB1: Homo sapiens, EAW62806; HsLCB2: H. sapiens, NP_004854; MmLCB2: Mus muscalis, NP_035609; DmLCB2: Drosophila melanogaster, BAA83721; CeLCB2: Caenorhabditis elegans, Q20375; DrLCB2, Danio rerio, NP_001108213; OsLCB2: Oryza sativa, BAD88168.1; AtLCB2: Arabidopsis thaliana, NP_001031932.1; DdLCB2: Dictyostelium discodium, XP_635115. Joint Genome Institute accession numbers: NvLCB2: Nematostella vectensis, 241814; MbLCB2: Monosiga brevicollis, 34401; PtLCB2: Populus trichocarpa, 834365; OtLCB2: Ostreococcus tauri, 16411; TpLCB2: T. pseudonana, 255691; PrLCB2: P. ramorum, 71166; NgLCB2: Naegleria gruberi, 82916; SpSPT: S. paucimobilis, Q93UV0_PSEPA; SmSPT: S. multivorum, A7BFV6_9SPHI; SsSPT: Sphingobacterium spiritivorum, A7BFV7_9SPHI; BsSPT: Bacteriovorax stolpii, A7BFV8_9DELT. Universal Protein Resource accession numbers: ScLCB2: S. cerevisiae, P40970; CnLCB2: Cryptococcus neoformans, J9VYF7; SpLCB2: Schizosaccharomyces pombe, Q09925; AnLCB2: Aspergillus nidulans, Q5BEC8; RoLCB2: Rhizopus oryzae, I1BXF5. Cyanidioschyzon merolae Genome Project accession number: CmLCB2: Cyanidioschyzon merolae, CMJ240C. ToxoDB accession numbers: TgSPT1: T. gondii, TGME49_090980; TgSPT2, TGME49_090970. GeneDB accession numbers: PfSPT: P. falciparum, PF14_0155; PvSPT, P. vivax; CmSPT: C. muris, B6ACS8_CRYMR; TbLCB2: Trypanosoma brucei, Tb927.10.4050; LmLCB2: Leishmania major, LmjF35.0320. Sanger Institute accession number: EtSPT: E. tenella, dev_EIMER_contig_00020813.