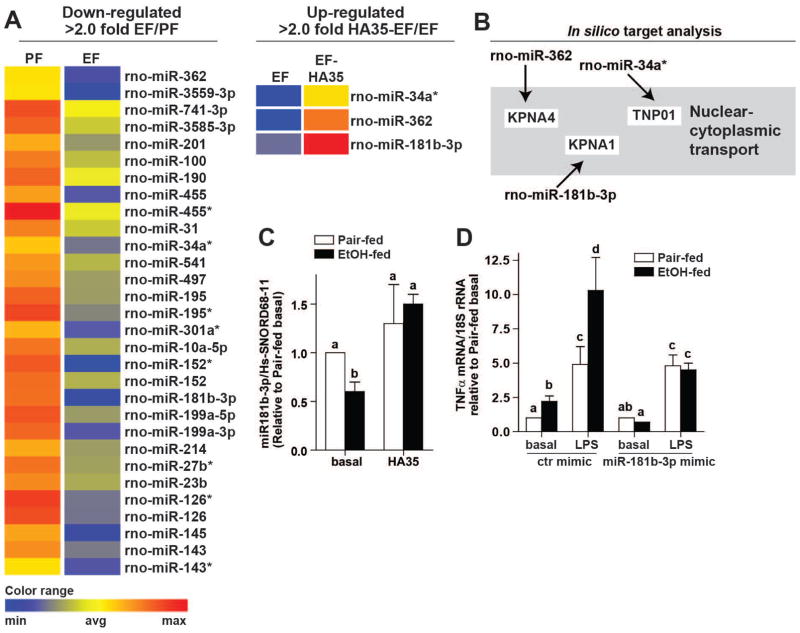
Figure 2. Next Generation Sequencing identifies miR181b-3p as a negative regulator of LPS-stimulated TNFα in Kupffer cells from ethanol-fed rats.
(A) miRNA isolated from Kupffer cells from ethanol- and pair-fed rats treated or not with 100μg/ml HA35 for 5 h was analyzed by NGS. Heat maps illustrate the changes in expression for the 30 miRNAs identified whose expression was decreased by more than 2-fold by ethanol feeding. Of these thirty miRNA, heat maps are shown for the 3 miRNA whose expression was restored by treatment with HA35. Fold changes are shown in Supplemental Tables 2 and 3. (B) Cartoon illustrates an in silico gene targeting analysis identifying nuclear-cytoplasmic shuttling as a common target of the three reciprocally regulated miRNA. (C) Kupffer cells isolated from ethanol- and pair-fed rats were treated or not with HA35 for 5 h. Expression of miR181b-3p was measured by qRT-PCR and normalized to Hs-SNORD68-11. (D) Kupffer cells were nucleofected with either control miRNA mimic or miR181b-3p mimic. 18hr post-nucleofection, Kupffer cells were challenged with 10ng/ml LPS for 1 hr and expression of TNFα mRNA measured by qRT-PCR and normalized to 18S rRNA. n=4.