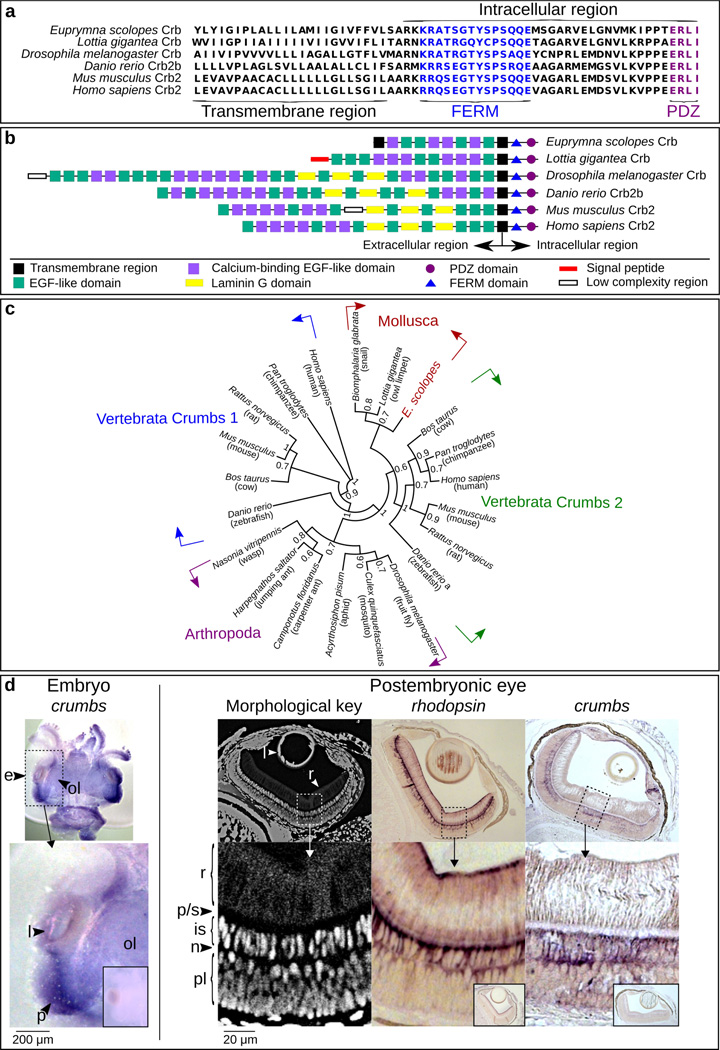
Figure 2.
Evidence for the presence of the Crumbs protein in E. scolopes. (a) Alignment of the C-terminus of EsCrumbs indicating the transmembrane region and the conserved FERM and PDZ domains within the intracellular region (see also Richard et al 2006). (b) Domain structures throughout the full EsCrumbs and the Crumbs proteins of other organisms as predicted by SMART (http://smart.embl-heidelberg.de/). (c) Phylogenetic position of EsCrumbs relative to those of other animal species. Supports are from 500 bootstrapped replicates. (d) Localization of es-crumbs during early development by in situ hybridization with either whole-mount or histological samples. Left: A stage 22 embryo (~1/2 through embryogenesis at 14 days) showing es-crumbs localized to the region of the eye (e). Features of the eye include the lens (l), optic lobe (ol), and region of development of the photoreceptors (p), indicated for reference. Right: Morphological key for the cephalopod eye, showing the rhabdoms (r), proximal region/supporting cell layer (p/s), inner segments (is), photoreceptor nuclei (n), and plexiform layer (pl). For comparative labeling in the eye, es-rhodopsin expression occurred at the extreme ends of the rhabdoms similar to the protein localization in another squid species (Doryteuthis pealeii; Kingston et al. 2015), but also extended into the inner segments. Labeling of es-crumbs occurred within the inner segments. The sense controls for the ISH experiments involving the embryo and postembryonic eye are indicated in the small boxed regions. Lens objective: 10×.