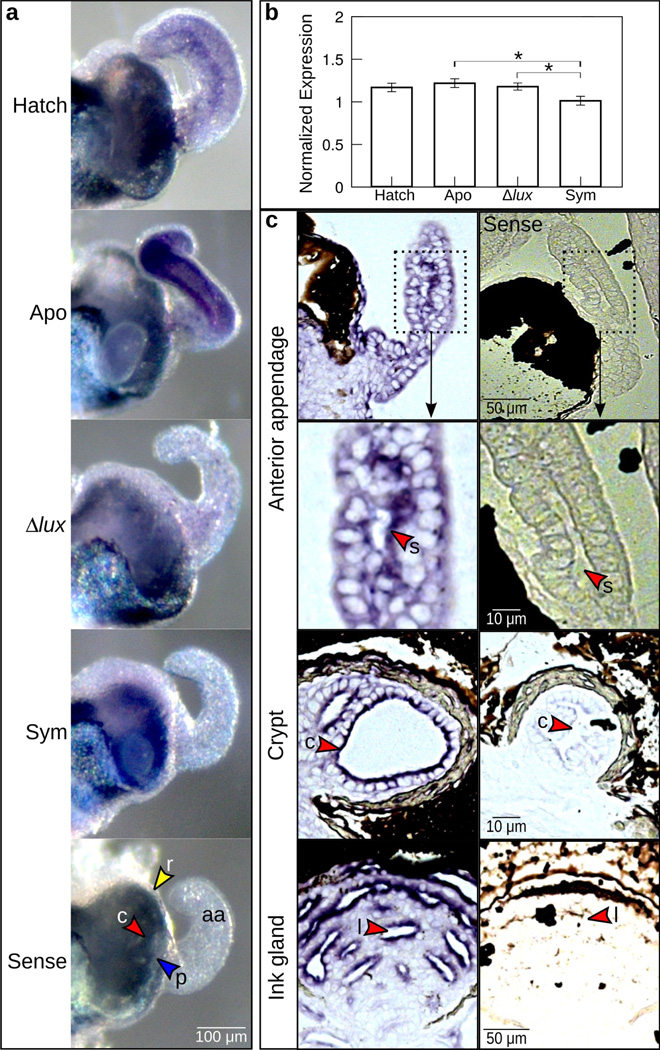
Figure 3.
Expression of es-crumbs throughout the light organ. (a) Whole light organs showing es-crumbs expression by whole-mount in situ hybridization. Conditions consisted of newly hatched juveniles (‘Hatch’) and 24-h juveniles aposymbiotic with no V. fischeri (‘Apo’), symbiotic with Δlux V. fischeri (‘Δlux’), and symbiotic with wild-type V. fischeri (‘Sym’). Tissues of the light organ showing es-crumbs signal included the anterior appendages (aa), ciliated ridges (r, yellow arrowheads), pores (p, blue arrowheads), or crypts (c, red arrowheads), and these features are shown in the ‘sense’ (control) image. (b) Regulation of es-crumbs by qRT-PCR in response to the different symbiotic conditions. Expression of es-crumbs was normalized to the geometric mean of the expression of two housekeeping genes, ribosomal 40s and serine HMT. Data consist of five biological replicates each with three technical replicates pooled per condition. Error bars are standard error of the mean (*, 0.01< P<0.05). (c) Histological sections of the light organ anterior appendage sinus (s) and crypt (c), and ink gland lumen (l) showing escrumbs localization by ISH (see red arrowheads). The images shown are from an animal colonized with Δlux V. fischeri after 24 h. The sense controls are shown along the right side.