Figure 8.
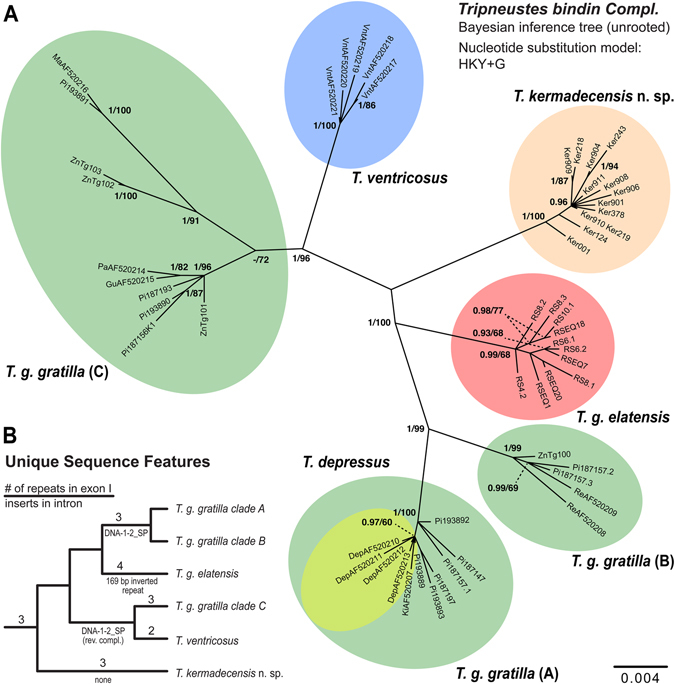
Phylogenetic tree reconstruction of Tripneustes bindin sequences. (A) Unrooted BI tree of 1256 bp long Tripneustes bindin sequences including exons 1 and 2 (excluding the glycine-rich repeat in exon 1; see main text for details) and parts of the intron (excluding the 169 bp region identified as transposon, respectively inverted repeat; see main text for details). The tree reconstruction is based on 54 unique haplotypes representing all extant species of Tripneustes. Supporting values (>0.9 posterior probabilities and >60% ML bootstrap values) are shown above the nodes. Color code as in Fig. 5. T. g. gratilla clades (A–C) are discussed in the text. Details on the sequences used for this tree are given in the Supplementary Tables S1 and S2. (B) Unique sequence features mapped on the bindin tree topology (position of root inferred from COI tree in Fig. 5). Numbers above branches represent number of gylcine rich repeats, names below branches provide information on the nature of insert at the 5′ end of the bindin intron (see text for details). Note that the transposon DNA-1-2_SP is inserted in normal orientation in T. g. gratilla clades A and B, but as reverse complement in T. ventricosus and T. g. gratilla clade C.
