Figure 1.
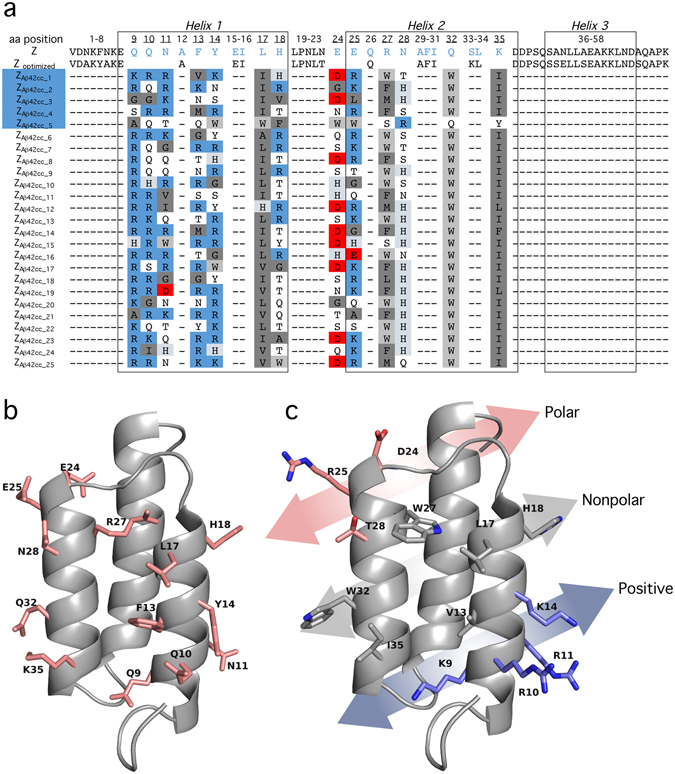
Sequence and cartoon representation of Affibody molecules selected as Aβ42cc protofibril binders. (a) Sequences of the top 25 selected Affibody molecules. The underlined amino acid positions are randomized in the phage display selection. The helical secondary structures are represented in boxes. The Affibody molecules chosen for further characterization are highlighted in blue. From 744 randomly picked clones, the sequence of ZAβ42cc_1 was identified eight times, ZAβ42cc_2 and ZAβ42cc_3 one time, ZAβ42cc_4 eight times and ZAβ42cc_5 three times. (b) The original Z-domain scaffold. Side chains of residues that are subjected to variation in the phage library are indicated. (c) Representation of side chains that are mutated in binder ZA β 42cc_1. The chemical properties of the side chains of the selected binders group into three chemically distinct regions of the surface, which identifies a ‘positive-nonpolar-polar’ recognition surface pattern.
