Figure 1.
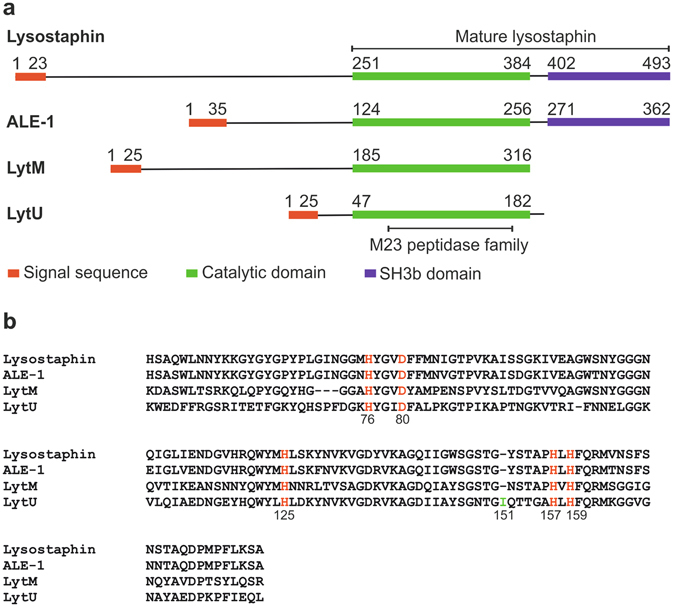
Comparison of M23 endopeptidase amino acid sequences. (a) Comparison of full-length lysostaphin, ALE-1, LytM, and LytU. N-termini of all proteins begin with a signal sequence that directs them to the cellular membrane and outward of the cytoplasm. The enzymes have unique N-domains (those for ALE-1 and lysostaphin are analogous, consisting of tandem 13 amino acid length repeats), for which functions remain unknown. The N-domain may need to be cleaved off for the enzymatic activity of catalytic domain (as for LytM, the activating in vivo enzyme is unknown20), need not be cleaved off (ALE-117), or the enzyme may increase activity upon cleavage (4.5-fold for lysostaphin21). Lysostaphin and ALE-1 possess, while autolytic LytM lacks SH3b domain downstream their catalytic domains to target extracellular substrates20, 42, 43. Structural predictions for LytU were made using SignalP44, HMMTOP45 and MODELLER46. Signal sequences are marked as indicated in the UniProt database, except for LytU sequence, which was predicted using SignalP44. (b) Alignment of the catalytic domains with ClustalW47. Conserved residues involved in zinc ion coordination (H76, D80 and H159) and catalytic (H125 and H157) are marked in red. The LytU unique insertion is highlighted in green and can be lysine in some S. aureus species.
