Figure 8.
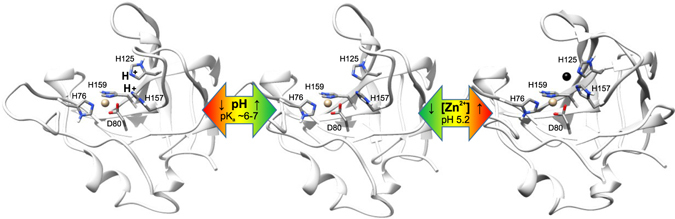
The constraints of LytU catalysis by inactivation with pH and zinc ions. Three structures of LytU catalytic domain are presented to the viewer peering through substrate binding groove (upper left in all three structures) with the site of catalysis at the proximal end. The key conserved residues are indicated. H76, D80 and H159 coordinate zinc (tan sphere) mandatory for substrate cleavage. Catalytic H157 and H125 reside nearby and are subjects to one of three fates. The block arrows indicate the driving forces in respective transitions and their gradient colour emphasizes positive (green) or negative (red) effect on LytU catalytic potential. The enzyme is in an active form (centre) and rapidly loses its activity (left) with decreasing pH (Supplementary Fig. S5). This is consistent with the protonation of H125 and H157. On the other hand, a second zinc ion (black sphere, right) may also bind LytU with a submicromolar dissociation constant (Supplementary Figs S6, S7 and Supplementary Table S2) and it is coordinated by Nε2 of the H125 and H157 (Supplementary Fig. S6). Consequently, this binding is catalytically inhibitory. Supplementary Fig. S10 provides a close-up look at the tautomeric forms of histidines.
