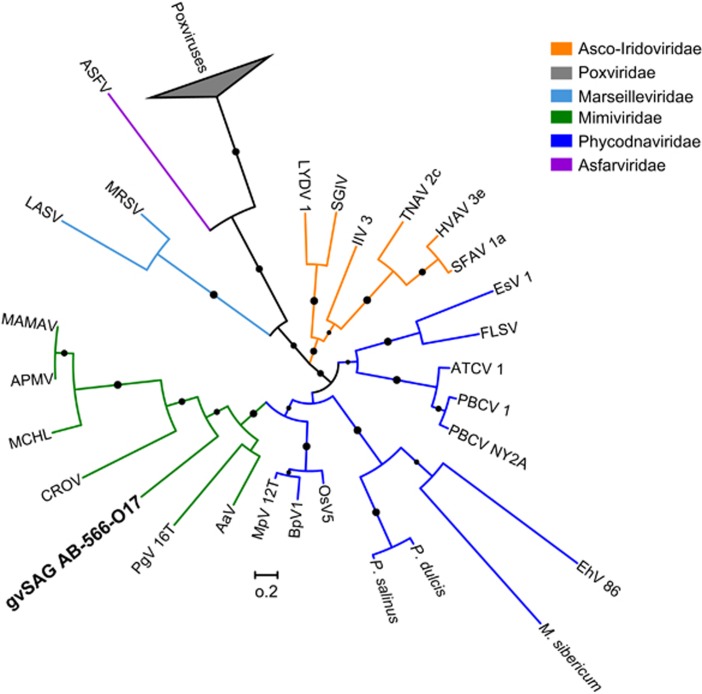
Figure 2.
Maximum likelihood phylogenetic tree of gvSAG-566-O17 and other NCLDV members constructed from A32 virion packaging ATPase, a NCLDV core gene. Support values >50% are shown at the nodes as black circles. NCBI gi numbers: MAMAV, Mamavirus (351737587); APMV, Acanthamoeba polyphaga Mimivirus (311977822); MCHL, Megavirus chilensis (363539780), CROV, Cafeteria roenbergensis virus (310831328); PgV, Phaeocystis globosa virus (508181864); AaV, Aureococcus anophagefferens virus (672551239); MpV, Micromonas pusilla virus (472342694); BpV, Bathycoccus prasinos. virus (313768078); OsV, Ostreococcus sp. virus (163955071); P. salinus, Pandoravirus salinus (531037319); P. dulcis, Pandoravirus dulcis (526119111); M. sibericum, Mollivirus sibericum (927594479); EhV, Emiliania huxleyi virus (73852542); PBCV, Paramecium bursaria Chlorella virus (340025835); ATCV, Acanthamoeba turfacea Chlorella virus (155371384); FLSV, Feldmannia sp. virus (197322436); EsV, Ectocarpus siliculosus virus (13242498); SFAV, Spodoptera frugiperda ascovirus (115298612); HVAV, Heliothis virescens ascovirus (134287264); TNAV, Trichoplusia ni ascovirus (116326781); IIV, Invertebrate iridescent virus (109287966); SGIV, Singapore grouper iridovirus (56692771); LYDV, Lymphocystis disease virus (13358448); ASFV, African swine fever virus (9628186); MRSV, Marseillevirus (284504185); LASV, Lausannevirus (327409704).