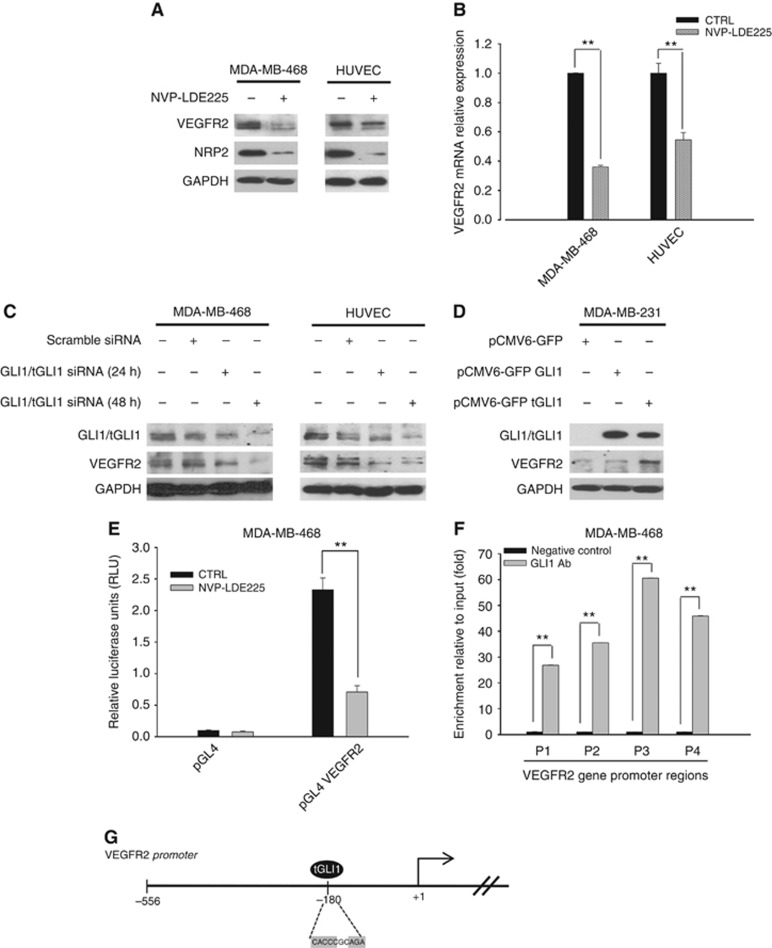
Figure 4.
GLI1 regulates VEGFR2 expression. (A) Western blot analysis of protein expression in MDA-MB-468 and HUVEC cells treated with NVP-LDE225 (2.5 μM). (B) Relative expression of sVEGFR2 mRNA in MDA-MB-468 and HUVEC cells treated with NVP-LDE225 (2.5 μM), as performed by real-time RT–PCR (qRT-PCR) analysis. Data were calculated with mean cycle threshold (CT) values, normalised to endogenous control. Data represent the mean (±s.d.) of three independent experiments, each performed in triplicate. (C) Western blot analysis of protein expression in MDA-MB-468 and HUVEC cells, 24 and 48 h after transfection with scramble or GLI1 siRNA pool (50 nmol l−1) using DharmaFECT 1 Transfection Reagent in DMEM. (D) Western blot analysis of protein expression in MDA-MB-231 cells, 24 h after transfection with either pCMV6-GFP empty vector, pCMV6-GFP GLI1 or pCMV6-GFP tGLI1 plasmids using lipofectamine 2000 in DMEM. (E) Relative luciferase units in MDA-MB-468 cells transfected with the empty pGL4 plasmid or the pGL4 plasmid containing 500 bp fragment of VEGFR2 promoter, and treated with NVP-LDE225 5 μM for 24 h after transfection. Luciferase activity was determined 48 h after transfection. Results were the average of three independent experiments. (F) Chromatin immunoprecipitation (ChIP) assay in MDA-MB-468 cells by using a GLI1 antibody and primers specific for the VEGFR2 promoter. Results were reported as fold change compared to negative control (no antibody); results were the average of three independent experiments. Bars, s.d. Asterisks indicate statistical significance, as determined by the Student t-test (**P<0.01). (G) Graphical representation of VEGFR2 proximal promoter region, containing the binding site of tGLI1 indicated by oval.