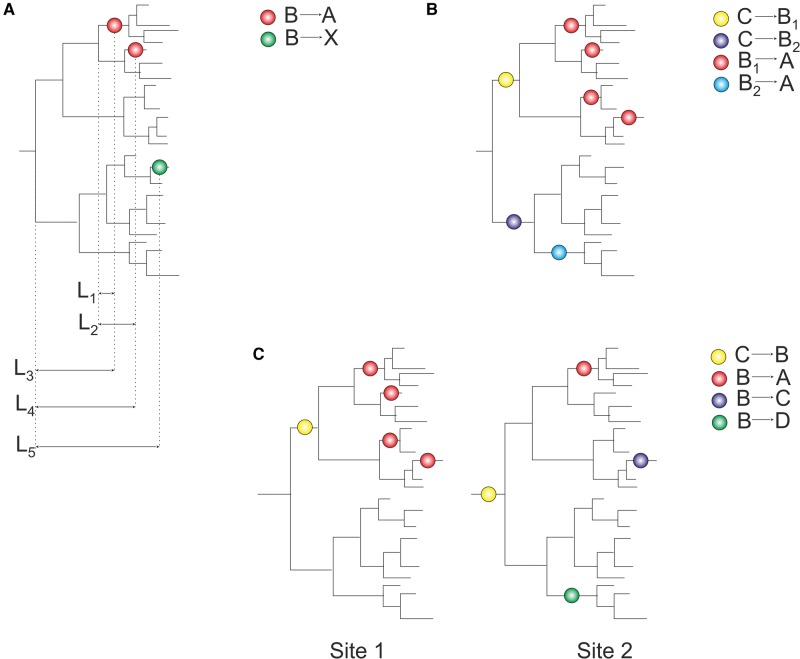
Fig. 1.—
Inference of phylogenetic distances between parallel and divergent substitutions. Dots represent substitutions mapped to nodes of a phylogenetic tree. (A) For each pair of amino acids (B, A) at a particular amino acid site, we consider the distances between all parallel B→A substitutions (L1 + L2), and distances between all divergent substitutions B→A and B→X (L3 + L5, L4 + L5), where X is any amino acid other than A and B. (B) The B1→A substitution is more frequent than the B2→A substitution, leading to an excess of homoplasies at small phylogenetic distances when parallel and convergent substitutions are pooled together. (C) Pooling of sites with different properties may also lead to an excess of homoplasies at small phylogenetic distances (see text).