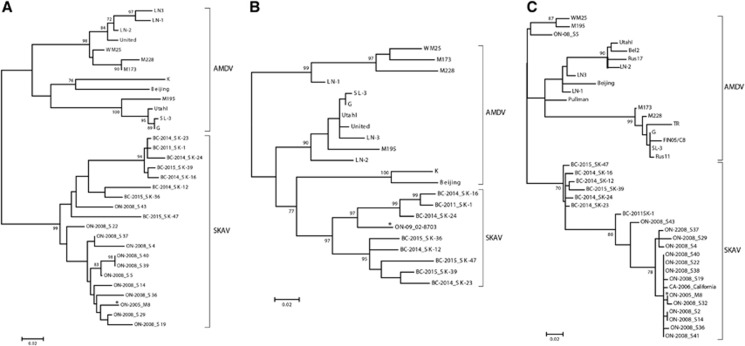
Figure 5.
Phylogenetic trees of AMDVs and SKAVs constructed with partial nucleotide sequences of the NS1 (A and B) and VP2 (C) regions. Trees have been constructed with alignments based on regions between nucleotides 602–922 (A), 1229–1672 (B), and 3018–3227 (C) of the AMDV-G complete genome (accession number JN040434). The evolutionary histories were inferred using the maximum-likelihood method19 based on the HKY30 (NS1) and TN9331 (VP2) models. AMDV and SKAV species are indicated by square parenthesis and the name of each strain is preceded by the location (British Columbia, BC; Ontario, ON; California, CA) and year of collection. SKAV sequences identified in mink are labeled with an asterisk.