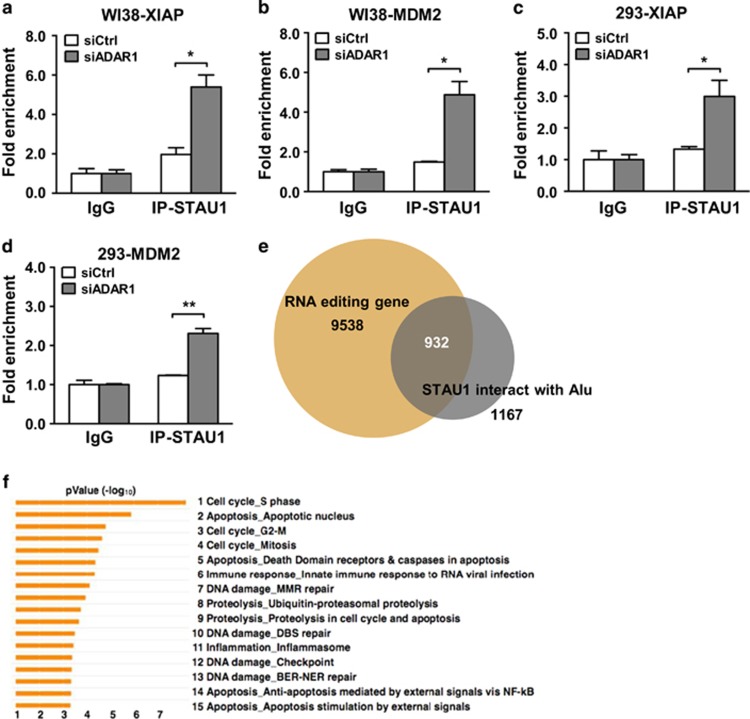
Figure 5.
Competitive occupancy of target 3′ UTR by ADAR1 and the RNA transport regulator STAU1. (a–d) Extent of STAU1 association with target transcripts was assessed by RIP assay. Lysates prepared from control (siCtrl) and ADAR1-depleted (siADAR1) cells (WI38 in a and b, 293 in c and d) were immunoprecipitated by the control IgG or STAU1 (IP-STAU1) antibodies. Bound RNA corresponding to the indicated targets (XIAP in a and c, MDM2 in b and d) was quantitatively determined by real-time RT-PCR. Data presented were normalized to the values of IgG in the control group and shown as means±S.D. (NS, not significant or P>0.05; *P<0.05; **P<0.01; ***P<0.001.) (e) The Venn diagram shows the degree of overlap between annotated RNA editing genes in the DARNED database and the known STAU1-interacting transcripts reported previously.34 (f) The 932 targets shared by the above two gene sets are enriched in distinct cell cycle and apoptosis processes, as revealed by bioinformatics analysis using MetaCore (P-value<0.05 and false discovery rate<0.05)