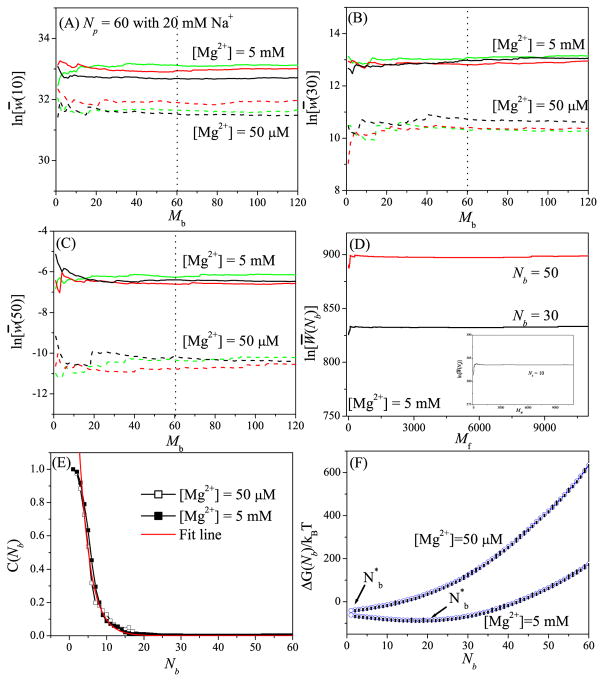
Figure 1.
Test results for the robustness of the Monte Carlo sampling in the “insertion-deletion” algorithm using a 60- nt RNA helix as the test case. (A) i = N/6, (B) i = N/2, and (C) i = 5N/6 show the results for the deletion process. For each of the three different sets (red, black, and green) for the N-ion (N = 60 here) distribution (generated through the insertion process), we run Mb Monte Carlo samples for the deletion process to generate an ensemble of the distributions for the i − 1 ions. Based on the distribution of the i − 1 ions, the statistical weight w(i) for the i-th ion is computed. (D) and the inset in (D) show the results for the Monte Carlo sampling in the insertion process. Here we show the results for Nb = N/6 (inset), Nb = N/2 (black line), and Nb = 5N/6 (red line). (E) Correlation (C(Nb)) between the statistical weights before and after the deletion step. The characteristic correlation-specific TB ion number is found to be 3 for this system. (F) The free energy ΔG(Nb) as a function of Nb. The error bar shows the fluctuation for the different Monte Carlo samples of the TB ion distribution.