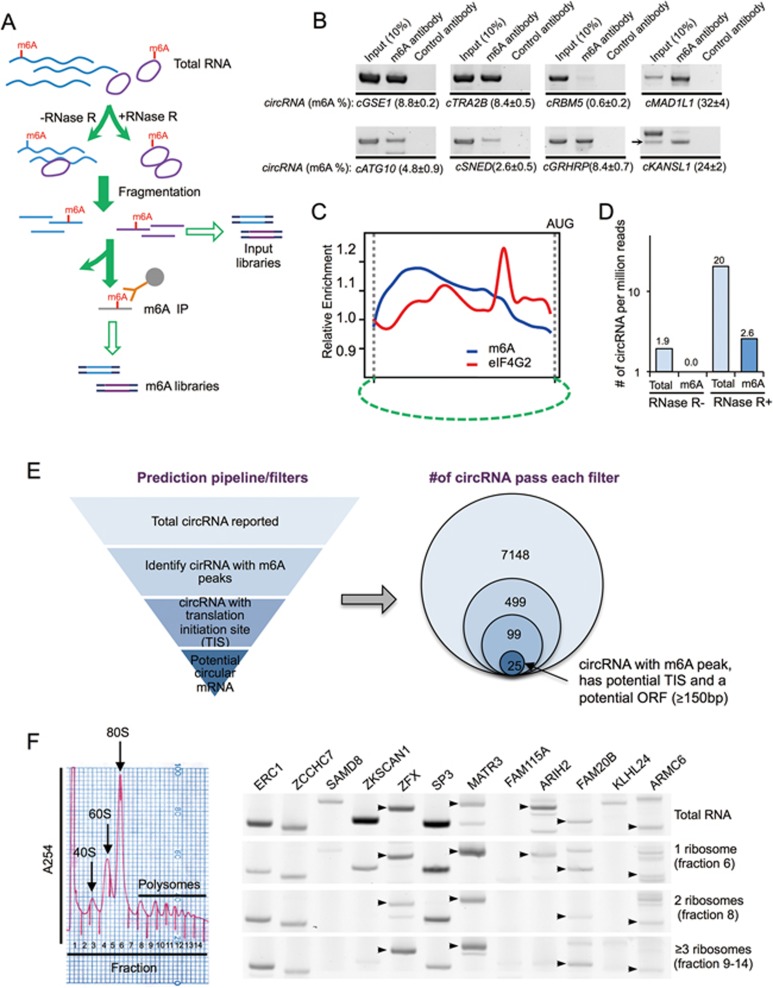
Figure 4.
Transcriptome-wide sequencing of m6A-modified circRNAs and predictive identification of endogenous circular mRNAs. (A) Schematic diagram of circRNA-m6A-seq protocol. (B) Validation of m6A-modified circRNAs in immunoprecipitated samples using m6A antibody or control antibody. Arrows indicate predicted circRNA size in the lanes with multiple bands, input (10%) indicates 10% of total input RNAs were used for RT-PCR, % m6A indicates percentage of m6A modification in target circRNAs (m6A antibody/(10× input (10%)). (C) Positional distribution of relative density for m6A and eIF4G2 binding site in circRNAs as compared to the respective control samples. The putative start codon was used as arbitrary marker to align the plot. When multiple AUG sites are presented in the circRNAs, the AUG that generates the longest ORF is use. (D) Number of circRNA reads (i.e., back-splice junction reads) per million of total reads in circRNA-m6A-seq samples treated with or without RNase R. (E) Schematic diagram of translatable circRNA prediction pipeline. Left, computational filters sequentially applied to identify circRNAs that contain m6A site, translation initiation site (TIS) and an ORF with sufficient length. The circRNA, m6A and TIS-sequencing data are from published results (see Materials and Methods section). Right, the numbers of circRNAs passing each filters. (F) Detection of predicted circRNAs from various host genes in polysome fractions with RT-PCRs. In the lanes with multiple bands, the circRNAs with expected size are indicated with arrows. IP, immunoprecipitation.