Fig. 1. The small-molecule A18 specifically inhibits the binding of IL-17A to IL-17RA.
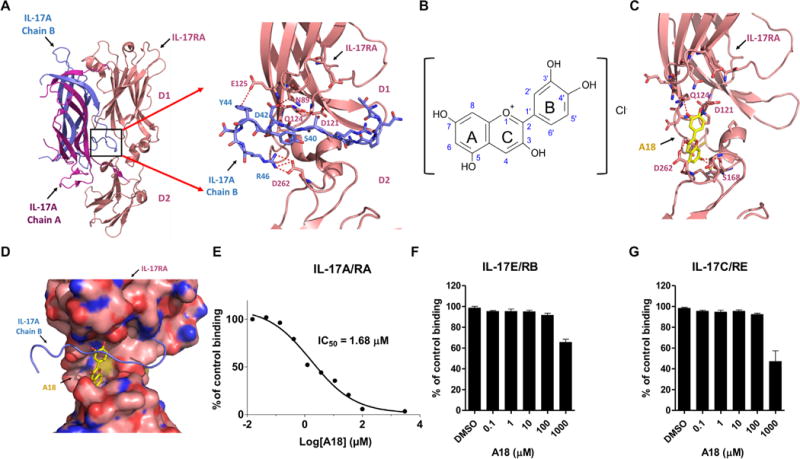
(A) Crystal structures of the IL-17A/IL-17RA complex [Protein Data Bank (PDB) 4HSA] and a magnified view of the binding interface (boxed region) indicating the deep ligand-binding pocket (defined in the text) with a cluster of hydrophilic residues highlighted at the interface. Chains A and B of IL-17A are shown in magenta and blue, respectively. The D1 and D2 domains of IL-17RA are shown in coral. (B) Chemical structure of A18. (C) Docking of A18 to the IL-17RA binding pocket. Red dashed lines indicate potential hydrogen bonds formed between A18 and critical amino acid residues in the docking pocket of IL-17RA. (D) Superimposition of the structures of IL-17A (blue) and A18 (yellow) onto the structure of the binding pocket of IL-17RA (pink). The binding mode of A18 was generated by docking. IL-17RA is shown in surface representation. (E to G) Analysis of the effects of the indicated concentrations of A18 on the extent of binding of IL-17RA–Fc to IL-17A (E), IL-17RB–Fc to IL-17E (F), and IL-17RE–Fc to IL-17C (G). The extent of binding inhibition by A18 was expressed as a percentage of the binding of the appropriate ligand to the indicated receptor in the absence of A18 [dimethyl sulfoxide (DMSO) control], which was set at 100%. Data are means ± SEM of three independent experiments.
