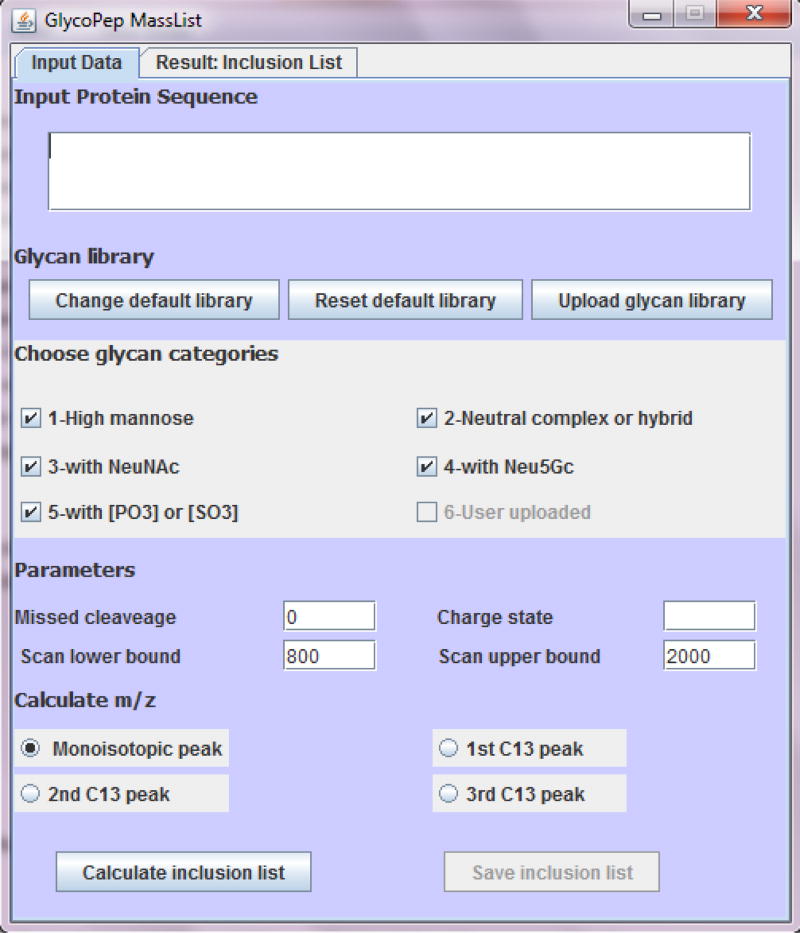
Fig. 1.
User interface for GlycoPep MassList. Users input the protein sequence, glycan library, charge state, mass range, number of missed cleavages, and select the mass (monoisotopic mass or 13C mass) to be calculated. In silico tryptic digestion is then performed on the protein and all the potential glycosylated peptides are reported by the software. The output is displayed under “Result: Inclusion List”. One or multiple groups of glycans can be selected, or custom glycan libraries can be uploaded, or a combination of manually input libraries and those in the database can be used