Figure 7.
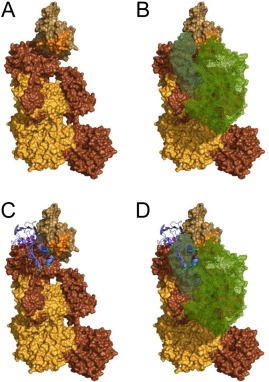
An experimentally constrained structural model for the C4b/Eap34 complex. An experimentally constrained model for the C4b/Eap34 complex was generated by the ClusPro server as described in the Materials and Methods section. (A) Surface rendering of C4b where each distinct polypeptide is shaded as follows: α′‐chain in brown, β‐chain in yellow‐orange, and γ‐chain in beige. The MIDAS‐acceptor site of the C345c domain, which mediates metal‐dependent binding of the vWF domain of C2 to C4b, is shaded orange. (B) Space filling model of the CP/LP C3 pro‐convertase, C4b2. C4b is colored as described in panel A, while the two regions of C2 (wire mesh) are colored with C2b in forest green and C2a in light green. (C) C4b/Eap34 structural model where C4b is colored as in panels A and B, while Eap34 is shown as a purple ribbon. The locations of key lysine residues in Eap34 are highlighted in magenta (Eap3) and cyan (Eap4), consistent with Figure 4(B). (D) Superposition of the structures shown in panels B and C. Note the similar locations of the C2b and Eap34 binding sites on C4b, and the potential for extensive steric clash if both proteins were present. This model is therefore in good agreement with the previously described ability of Eap34 to potently compete with C2 for C4b binding.8
