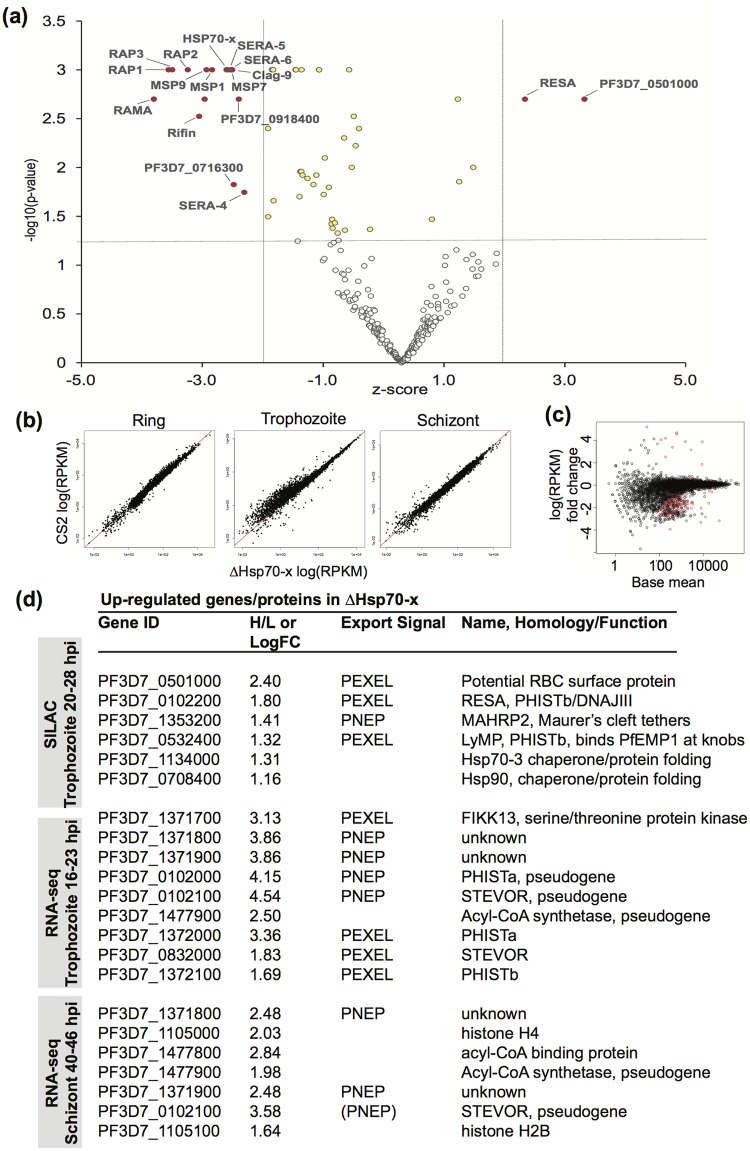
Fig 6. Transcriptional and translational changes in ΔHsp70-x indicate an up-regulation of some exported proteins.
(a) Mass spectrometry based SILAC sequencing of proteins indicates several exported proteins are over- and under-expressed in ΔHsp70-x relative to CS2. ΔHsp70-x and CS2 were differentially labeled with heavy (H) and light (L) isotopic forms of isoleucine respectively. Proteins with statistically insignificant Z-score ratios (X-axis) versus—log10 (P-value) (Y-axis) and therefore in ~ 1:1 ratio represent the base of the volcano plot. Proteins at higher levels in ΔHsp70-x only and which satisfy both statistical parameters are labeled in red in the upper right. Those up-regulated proteins that only satisfy one parameter are labeled in yellow. Proteins down-regulated in ΔHsp70-x are similarly coloured and shown in the upper left. (b) Correlation plots of RNA-seq transcripts between wild type and ΔHsp70-x ring, trophozoite and schizont stages show close correlation in most transcripts (close to line x = y). Most differential genes are downregulated in ΔHsp70-x trophozoites and schizonts (points furthest from line). (c) MA plot representing RNA-seq data. The change in expression (log 2 fold change) is plotted against the average of the normalized count values (base mean), of ΔHsp70-x and CS2 transcripts at the trophozoite stage. Red dots indicate differentially expressed genes in ΔHsp70-x. (d) Summary of increased proteins and transcripts by SILAC and RNA-seq. Numbers indicate heavy/light isoleucine ratio (H/L) in SILAC ordered by the ratio, or the log2 fold change in transcripts (LogFC) in RNA-seq data, ordered by significance which takes into account number of transcripts as well. Exported proteins are denoted as PEXEL or no PEXEL (PNEP) or otherwise left blank. Known homology to protein families or potential function identified in final column, including annotated pseudogenes. Pseudogenes may produce a limited numbers of transcripts so small changes can look significant.