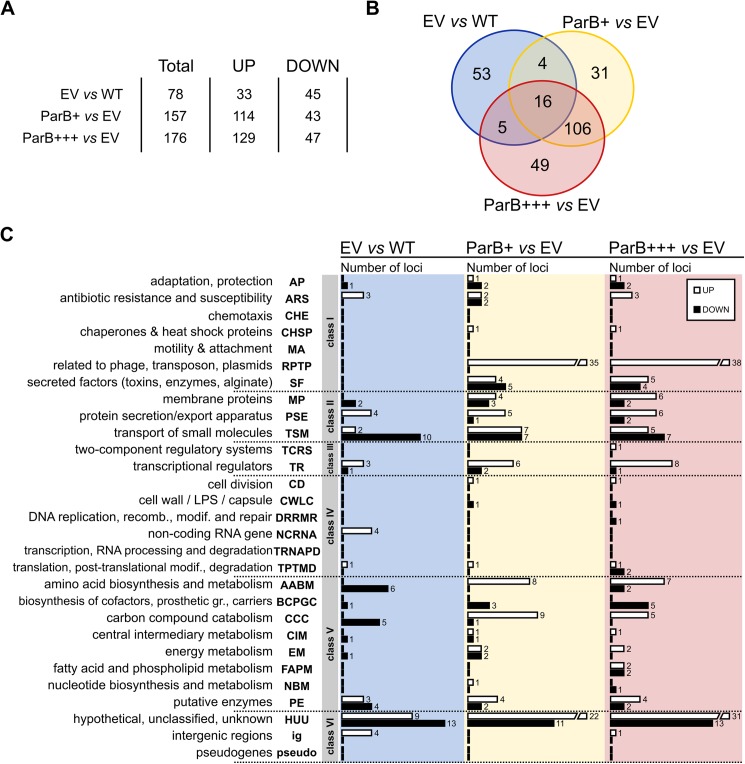
Fig 2. Transcriptome changes in response to ParB overproduction.
(A) Statistics of loci with significant expression change (FC<-2 or >2, p-value <0.05). RNA was isolated from PAO1161 cultures grown in L broth without Ara (WT), PAO1161 (pKGB8 araBADp) cultures grown under selection in L broth with 0.02% Ara (empty vector control, EV), PAO1161 (pKGB9 araBADp-parB) cultures grown under selection in L broth without Ara (mild ParB excess, ParB+) or with 0.02% Ara (higher ParB excess, ParB+++). (B) Venn diagram for sets of loci with significant expression change between EV vs WT, ParB+ vs EV and ParB+++ vs EV. (C) Classification of loci with altered expression according to PseudoCAP categories [57]. When a gene was assigned to multiple categories, one category was arbitrarily selected (S2 Table). The PseudoCAP categories were grouped into six classes as marked. White and black bars correspond to the numbers of respectively, upregulated and downregulated genes in a particular category.