Figure 2.
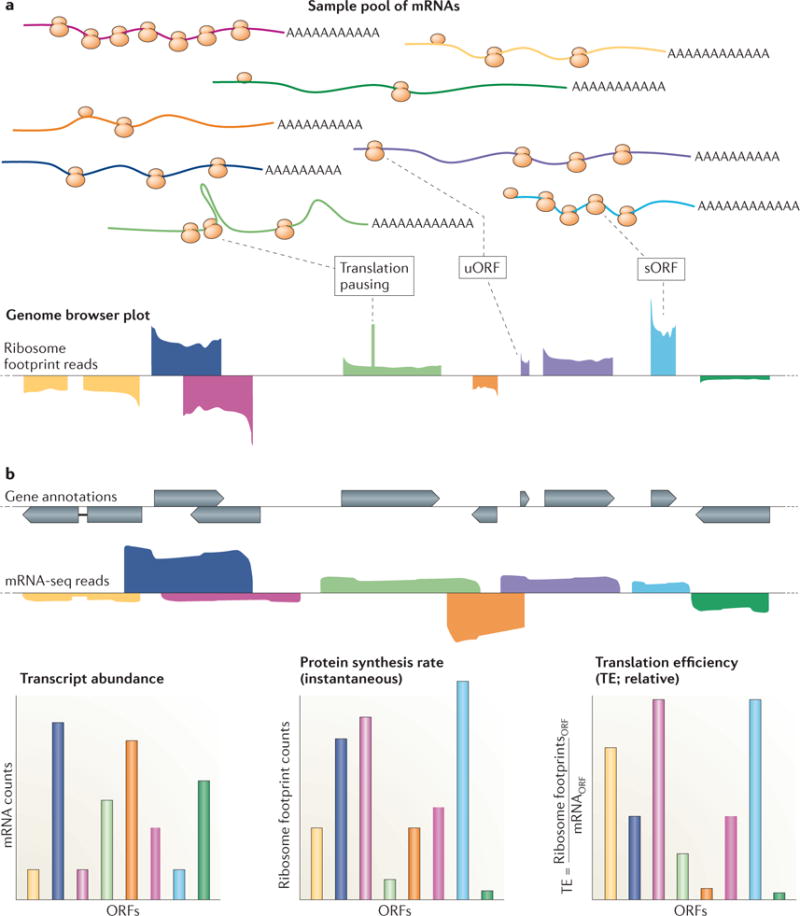
Qualitative and quantitative data provided by ribosome profiling. a) A diverse sample pool of mRNAs, distinguished by color, are shown together with a corresponding representative genome browser plot of ribosome profiling data derived from this pool. Note that ribosome profiling enables experimental determination of translated regions, including short Open Reading Frames (sORFs), which may be an important new source of cellular peptides, and upstream ORFs (uORFs), which are thought to be largely regulatory. Pausing during translation elongation may result in peaks in ribosome footprints within ORFs. b) Overlaid gene annotations and mRNA-seq data for the examples shown in (a). c) Examples of quantitative data derived from b). Note that transcript abundances may not correlate closely with the instantaneous protein synthesis rates. The collection of quantitative data for both transcript abundances and their protein synthesis rates enables inference of the relative translation efficiencies. These can vary over several orders of magnitude within a given organism in a given state. The translation efficiency can also change over time for a given mRNA, reflecting dynamic regulation at the level of translation.
