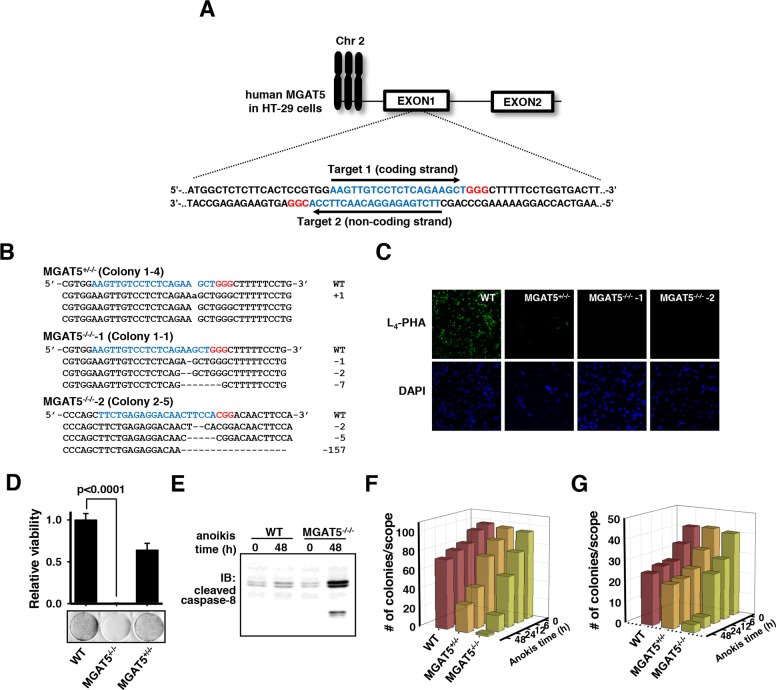
Figure 4. Validation of loss of anoikis resistance by MGAT5 gene knock-out in HT-29 cells using CRISPR/Cas9.
A. Two targets for sgRNA were selected at the first exon of the MGAT5 gene. Target and PAM sequences are noted in blue and red letters, respectively. B. Gene editing using CRISPR/Cas9 created one heterologous and two homologous MGAT5 knock-out colonies. The indel mutations with a frame shift were confirmed by Sanger sequencing. C. Knockout of MGAT5 was confirmed by immunofluorescence using biotin-labeled L4-PHA and FITC-labeled streptavidin. D. The viability tests were conducted by imposing anoikis stress for 48 hours and then counting the colonies formed on culture plates (n=3). E. Caspase-8 cleavage was monitored by immuoblot analysis. F-G. Changes in the colony-forming capabilities by MGAT5 knockout were monitored under anchorage-dependent (F) and -independent conditions (G). Cells were treated with anoikis stress for indicated times and embedded into Matrigels (F) and soft agars (G).