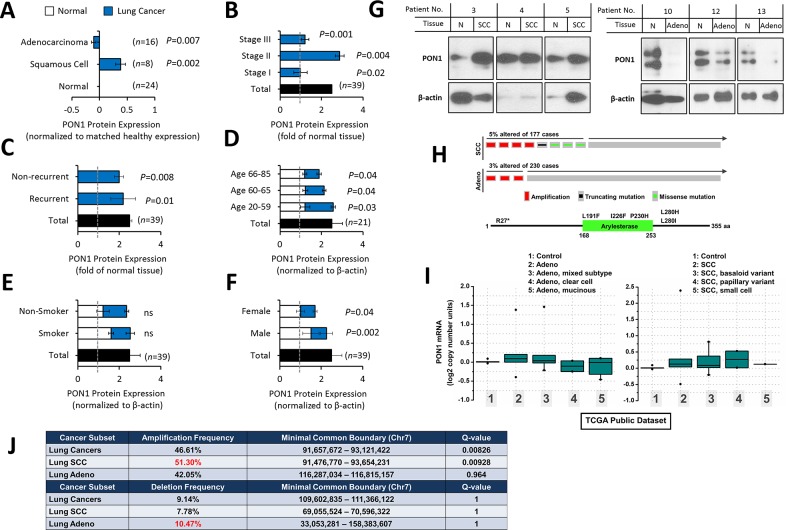
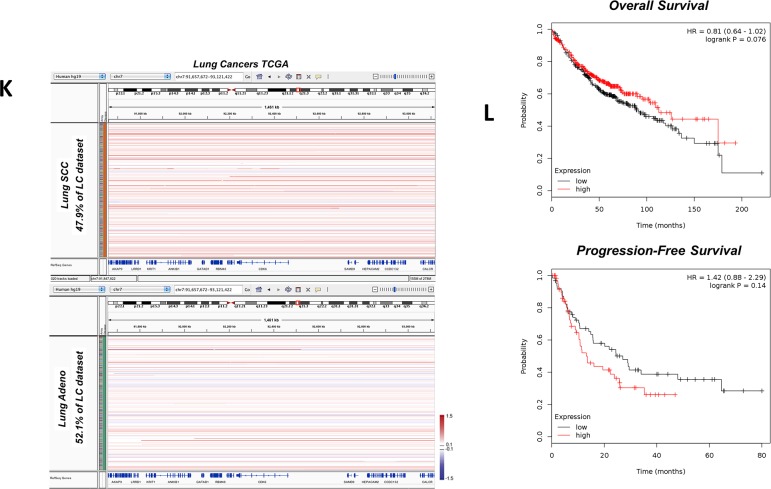
Figure 1. Variation of PON1 protein and gene expression between lung squamous cell carcinoma and lung adenocarcinoma patients.
(A) Densitometry analysis of lung cancer subtype tissue-based expression of PON1 protein from matched clinical pairs of lung squamous cell carcinoma and lung adenocarcinoma normalized to adjacent normal control. (B, C, D, E, F) Matched tissue samples were either summarized (total) or separated into different groups of: stage, recurrence, age, smoking history, and gender. Presented results were summarized from two biological replicates of the blotting experiment. (G) Representative blots of PON1 protein patient tissue expression selected from 39 matched cases. β-actin variations apparent in some blots were considered in calculating relative densitometry values for each blot using nonparametric Kruskal-Wallis statistical test. (H) Genetic alterations and mRNA expression changes (copy number) of PON1 in the TCGA dataset of lung cancer samples. PON1 genes are represented in rows, and individual cases or patients are represented as columns (upper panel). Higher magnification images are shown in the inset (lower panel). These oncoprints are based on data obtained from the cBio portal (http://www.cbioportal.org). (I) TCGA cohorts of lung cancer (lung squamous cell carcinoma and lung adenocarcinoma variants) gene expression study were analyzed using Oncomine (http://www.oncomine.org). (J) Characterization of amplification and deletion of the PON1 locus. Summary of both amplification and deletion frequency and boundary of alteration of the PON1 locus in the lung squamous cell carcinoma and lung adenocarcinoma tumor types. Data were obtained through query of the TCGA TumorScape database (Broad Institute). (K) Lung squamous cell carcinoma and lung adenocarcinoma patients exhibiting amplification at the PON1 locus were visualized using the Integrative Genomic Viewer (IGVv2.3; Broad Institute). Red bars represent amplification, blue bars represent the degree of deletion, and white bars represent no alteration. DNA copy number ratio is relative to a reference somatic DNA sample. It must be noted that the single patient representing a focal amplification/deletion at the PON1 locus (boxed in red in the profile graphs). (L) Correlation of PON1 expression with patient survival in lung adenocarcinoma. PON1 expression was stratified as high versus low against median expression. Graphs were plotted using kmPlotter (www.kmplot.com) with overall survival and progression-free survival within previously published datasets. Tissue protein samples from our cohorts were blotted three times and were biologically repeated three times