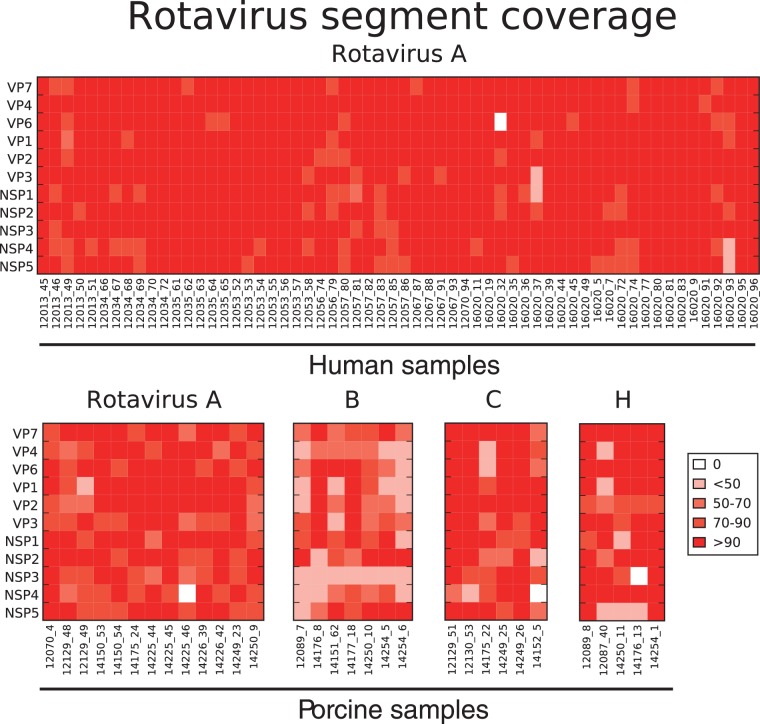
Figure 1.
Heat map of RV sequence length coverage by segment detected in all samples. The sequence length coverage for each segment of all assembled rotaviruses by deep sequencing was calculated and expressed as [(length of assembled contig in nt)/(full-length of that segment in nt)]×100. Colour code for %genome coverage is indicated in figure key ranging from low (pale orange) to high (dark red). The value of 0 indicates that contig sequence for that segment was not identified or did not pass the stringent quality control criteria including % reads mapped and contig length (see Materials and Methods). All RVA sequences detected in human samples were shown in the first panel, with each column representing a sample and each row showing each RV segment. Similarly, porcine RV samples were shown, each panel representing RVA, RVB, RVC, and RVH sequences with the segment names given vertically and sample IDs horizontally.