Figure 1.
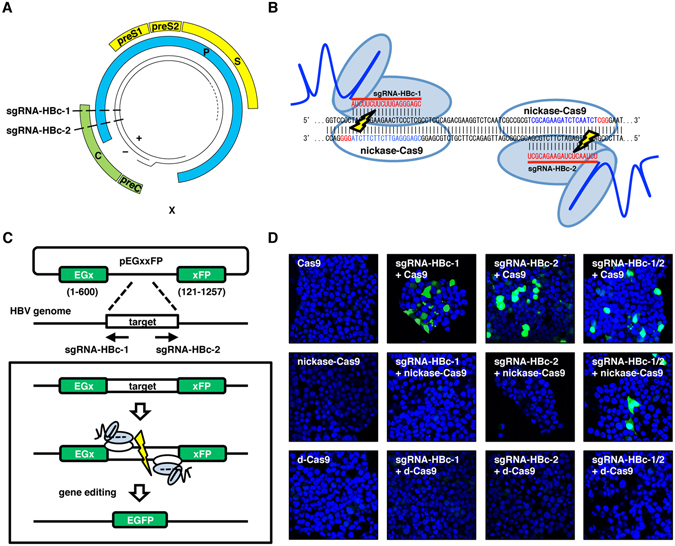
Cas9-based systems for gene editing and HBV-specific sgRNAs targeting sites for Cas9, nickase-Cas9 or d-Cas9. (A) Schematic diagram representation of the HBV genome. The four viral transcripts of the core are indicated: C, polymerase; P, surface; S and X, proteins. Regions targeted by sgRNAs are indicated. (B) The DNA sequences targeted by nickase-Cas9 with the pair sgRNA-HBc-1 and sgRNA-HBc-2 in the gene coding HBc protein. (C) Schematic of the EGxxFP assay performed to analyze the cleavage effects of Cas9, nickase-Cas9 and d-Cas9 expression with sgRNA-HBc-1 and sgRNA-HBc-2. A split EGxxFP plasmid is inserted with the targeted HBV sequence, and a recombination of EGFP is induced by DSBs on the targeted sequence. (D) Plasmids encoding Split EGxxFP possessing the HBc sequence, Cas9, nickase-Cas9, d-Cas9, and the sgRNAs were transfected into 293T cells. EGFP fluorescence was detected at 24 hr post-transduction.
