Figure 3. Pollen Tube Rupture Phenotype in en-RNAi Mutants and Interaction between ENODLs and FERONIA.
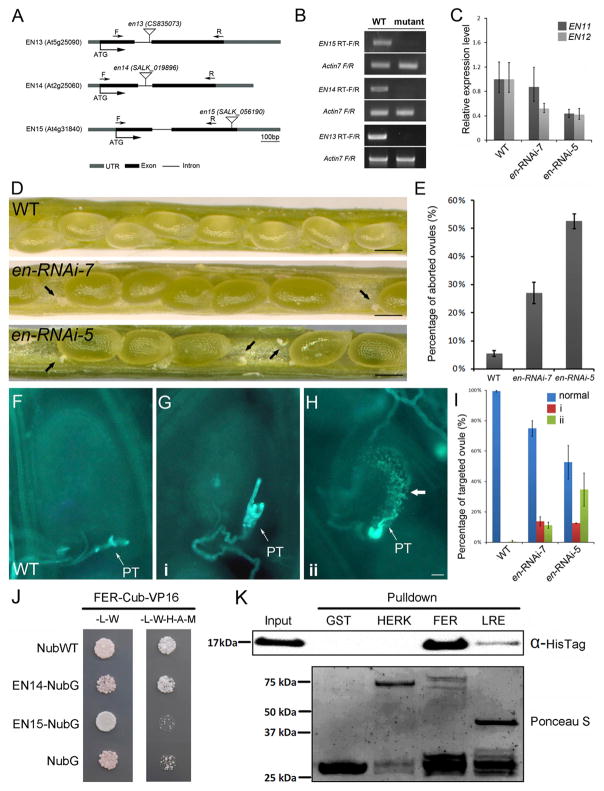
(A) Diagram showing T-DNA insertion sites in en13, en14, and en15 mutants.
(B) Semi-qRT-PCR analysis to show the expression levels of EN13, EN14, and EN15 in the corresponding SALK T-DNA mutant lines.
(C) Expression levels of EN11 and EN12 in en-RNAi mutants detected by qRT-PCR assays. The error bars represent the SD of three independent biological replicates of qRT-PCR experiments.
(D) Ovule abortion in en-RNAi siliques. Arrowheads indicate aborted ovules. Scale bars, 200 μm.
(E) Percentage of aborted ovules in two independent quintuple mutant lines compared with the wild-type (WT). Error bars are mean ± SD of three biological replicates; more than 50 siliques from each line were examined.
(F) Fertilized WT ovules. Aniline blue stains a single pollen tube (PT).
(G) Pollen tube failed to rupture and showed overgrowth in an en-RNAi mutant ovule (phenotype 1; i).
(H) Pollen tube failed to rupture and showed callose deposition in ovules of an en-RNAi mutant line (phenotype 2; ii). Arrow, callose deposition. Scale bar, 20 μm.
(I) Histogram to show quantification of both phenotypes. N > 120. Error bars are mean ± SD of three biological replicates; more than 50 siliques from each line were examined.
(J) ENODL14 strongly interacts with FERONIA (FER) in a split-ubiquitin yeast-two-hybrid assay. Cub-VP16, the C-terminal half of an ubiquitin fused with the transcription factor (LexA-VP16) to activate the reporter genes; NubWT, the N-terminal half of a wild-type ubiquitin; NubG, the N-terminal half of a mutated ubiquitin as a negative control; -L, medium without Leu; W, medium without Trp; H, medium without His; A, medium without Ade; M, medium without Met.
(K) ENODL14 strongly and specifically interacts with FER in a pull-down assay. ENODL14-HisTag (top) is pulled with GST-tagged extracellular domains (ECDs) of CrRLK-like receptor kinase FER, but not with its family member HERKULES (HERK). GST-LRE is weakly pulled down with ENODL14-His. The bottom panel shows GST pull-downs with Ponceau S staining.
See also Figure S4.