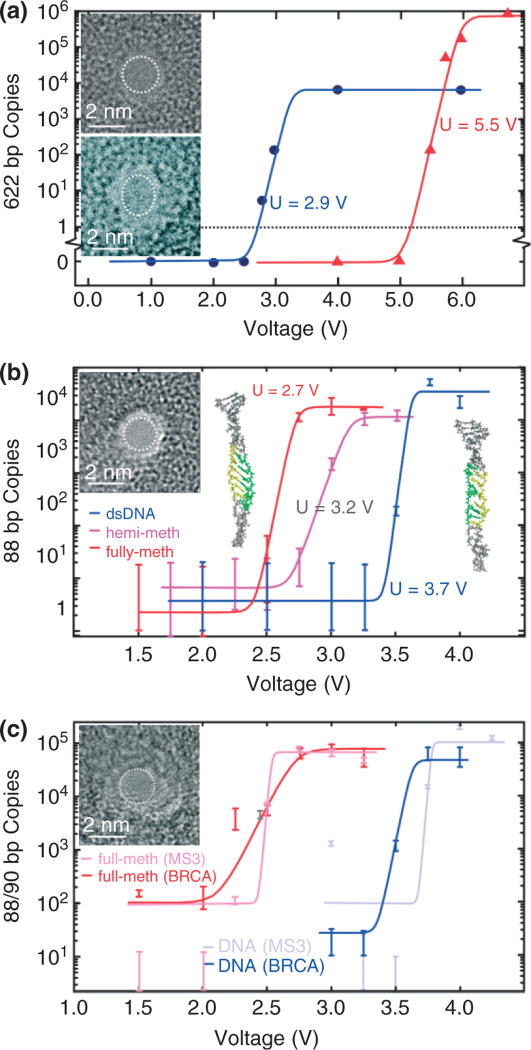
FIGURE 4.
Stretching DNA in a nanopore. (a) Quantitative PCR (qPCR) results obtained for 2-nm pores showing the copy number versus voltage. 622 base pair dsDNA permeates the 2-nm pore in 10-nm thick Si3N4 membrane (top inset) for V > 2.5 V and the 1.8 × 2.2 nm in 20-nm thick Si3N4 membrane (bottom inset) pore for V > 5 V. (b) qPCR results indicating the number of MS3 DNA copies that permeates through the 1.8 ± 0.2 nm pore shown in the inset as a function of the membrane voltage. Insets on the left and right are snapshots of methylated and unmethylated MS3 translocating through the 1.8-nm pore. Both DNA exhibit an ordered B-DNA form, but there is a significant degree of disorder for unmethylated DNA. The highlighted region of the strand shows the portion of the DNA where methylated cytosines are located. The same region is also highlighted in the unmethylated strand for comparison. (c) qPCR results indicating the number of MS3 and BRCA1 DNA copies that permeates through the 1.7 ± 0.2 nm pore shown in the inset as a function of the membrane voltage. Unmethylated MS3 and BRCA1 permeate the U > 3.8 V and U > 3.6 V, respectively while the threshold for fully methylated MS3 and BRCA1 is U > 2.5 V and U > 2.7 V, respectively.