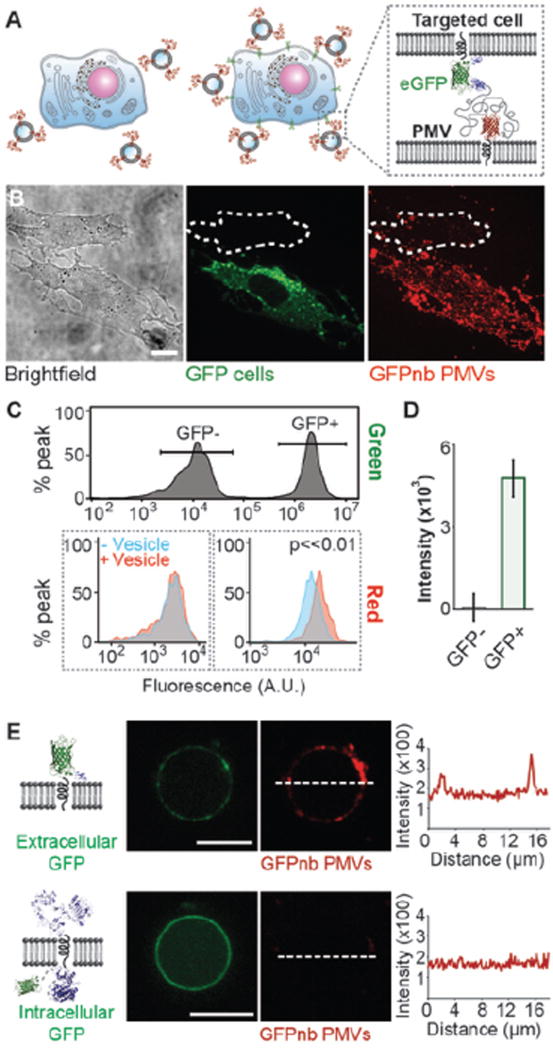
Figure 6. GFPnb-PMVs bind to eGFP expressing cells with high specificity.
A) Cartoon schematics showing competitive binding assay. Only cells that express GFP-tagged receptors on their surfaces are expected to recruit GFPnb-PMVs. B) The specificity GFPnb-PMV binding to cells was evaluated by co-culturing GFP negative and GFP positive cells in a 1:1 ratio (brightfield, green) and then exposing them simultaneously (in the same culture dish) to GFPnb-PMVs (red). GFP positive cells recruited GFPnb-PMVs in substantially greater quantities. The red fluorescent image is a maximum intensity Z projection. C) Flow cytometry analysis of GFPnb-PMV binding to the co-cultured cells. Top: The green fluorescence signals of GFP positive and negative cells were used to set separate gates to distinguish these two co-cultured populations of cells. Bottom: The overlay of recipient cell fluorescence with (red) and without (blue) GFPnb-PMVs. Only the GFP positive cells (right) have a clearly detectable fluorescence shift upon PMV binding. D) Mean fluorescence increase owing to GFPnb-PMV binding. E) GFPnb PMVs were used to demonstrate the preservation of transmembrane protein topology. GPMVs extracted from CHO cells transiently express transmembrane receptors with an extracellular GFP recruited GFPnb PMVs (top) while those from CHO cells transiently express EGFR with an intracellular GFP did not (bottom). The line scans show the intensity of mRFP signal from GFPnb PMVs. These images were taken under same camera setting and displayed using identical brightness and contrast for direct comparison. All scale bars, 10 µm.