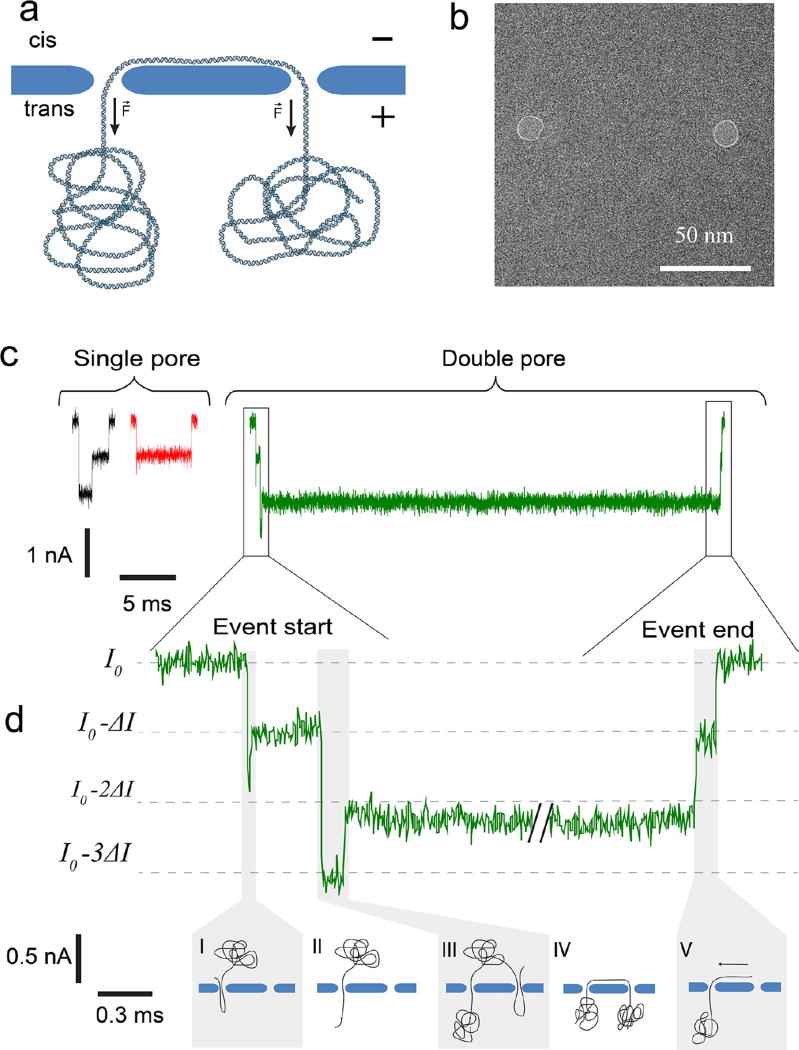
Figure 1.
Concept of trapping DNA in a double-nanopore system. (a) Side-view sketch of a single DNA molecule that is trapped in two nanopores. In a tug-of-war fashion, the forces in the two nanopores cancel out, thus arresting the translocation of the DNA. (b) TEM image of two 10 nm nanopores drilled in a freestanding SiN membrane, separated by 100 nm. (c) Typical examples of single-nanopore and double-nanopore events at a bias voltage of 300 mV, pore diameter of 15 nm, and the pore-to-pore distance of 550 nm. (d) Expanded view of the beginning and ending of the double-pore event. The DNA molecule enters the first nanopore in a folded conformation (I), subsequently traverses it in single-file fashion (II), whereupon a different part of the molecule is captured by the second pore (III) in a folded fashion. Finally, the DNA reaches the trapped state (IV), and eventually slides out (V).