Figure 3.
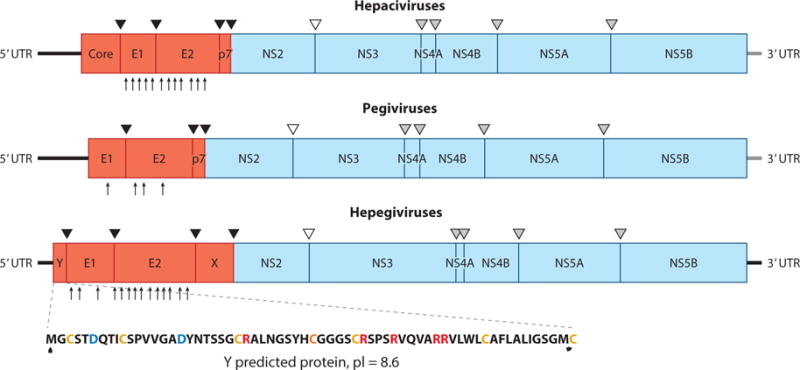
Genome organization and polyprotein cleavage map of hepaciviruses, pegiviruses, and hepegiviruses. The complete genome encodes a polyprotein that is co- and posttranslationally cleaved into individual viral proteins. The structural proteins include the core protein (C) and envelope glycoproteins (E1 and E2), and the nonstructural proteins are NS2, NS3, NS4A, NS4B, NS5A, and NS5B. Classical pegiviruses (human and simian pegiviruses) do not have a recognizable C protein. Structural proteins are cleaved by cellular signal peptidases (black triangles), and NS2-NS3 cleavage is accomplished by the NS2-NS3 autoprotease (empty triangle). The remaining nonstructural proteins are cleaved by the NS3-NS4A protease complex (gray triangles). Hepegiviruses encode a protein termed Y that is similar in location and properties to hepacivirus C. Glycosylation sites in E1 and E2 are shown by arrows at the bottom of the polyproteins for hepatitis C virus, GB virus C, and human hepegivirus (60, 114). Abbreviations: pI, protein isoelectric point; UTR, untranslated region.
