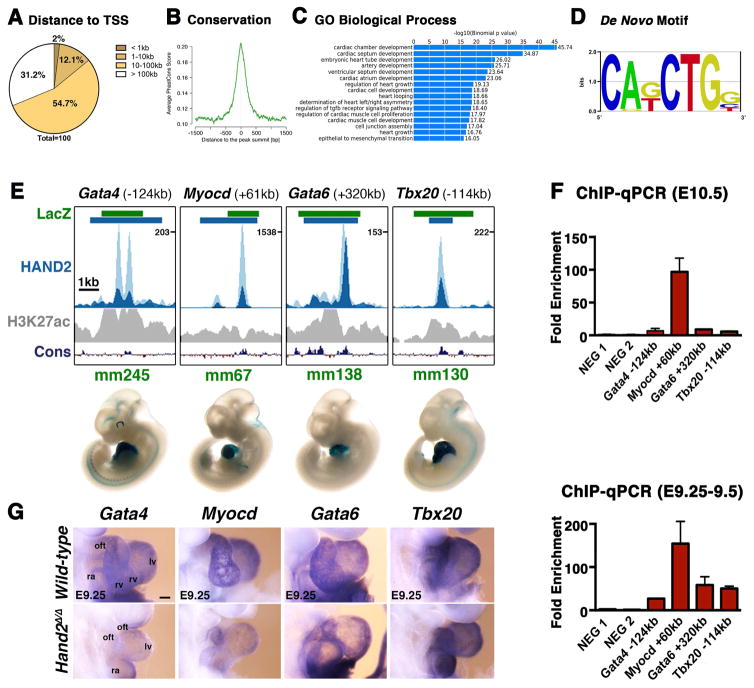
Figure 1. ChIP-Seq analysis using the Hand23xF allele identifies the HAND2 cistrome in mouse embryonic hearts.
(A, B) The majority of the genomic regions enriched in HAND2 chromatin complexes from mouse embryonic hearts at E10.5 map ≥10kb away from the closest transcription start site (TSS) and are evolutionarily conserved. (C) The top GO terms associated with HAND2 candidate targets reveal the preferential enrichment of genes functioning in cardiac development. (D) The consensus Ebox motif is most enriched by de novo motif discovery. (E) Selection of VISTA enhancers enriched in HAND2 chromatin complexes. Green intervals indicate the regions with enhancer activity (VISTA enhancer database); blue intervals highlight the regions enriched in HAND2 chromatin complexes (MACS peaks). Distances to the nearest TSS within the TAD are indicated on top. ChIP-Seq profiles of the two biological replicates (E10.5) are shown in light and dark blue, respectively. The H3K27ac ChIP-Seq profile for mouse hearts (E11.5) is shown in grey (Nord et al., 2013). The scheme at the bottom shows the placental mammal conservation (Cons) plot (PhyloP). Representative transgenic LacZ reporter embryos for VISTA enhancers associated to genes functioning in OFT and/or right ventricle development (Gata4, Myocd, Gata6 and Tbx20) are shown below. (F) ChIP-qPCR validation of the ChIP-Seq peaks (panel E) for mouse embryonic hearts at E10.5 (n=3 biological replicates) and E9.25 (n=2; mean ± SD). (G) Expression of the HAND2 targets Gata4, Myocd, Gata6 and Tbx20 in wild-type and Hand2-deficient (Hand2Δ/Δ) embryonic hearts (E9.0–9.5). Scale bars: 100μm. oft: outflow tract, ra: right atrium, rv: right ventricle, lv: left ventricle, avc: atrioventricular canal, la: left atrium. See also Figures S1–S3 and Tables S1–S4.