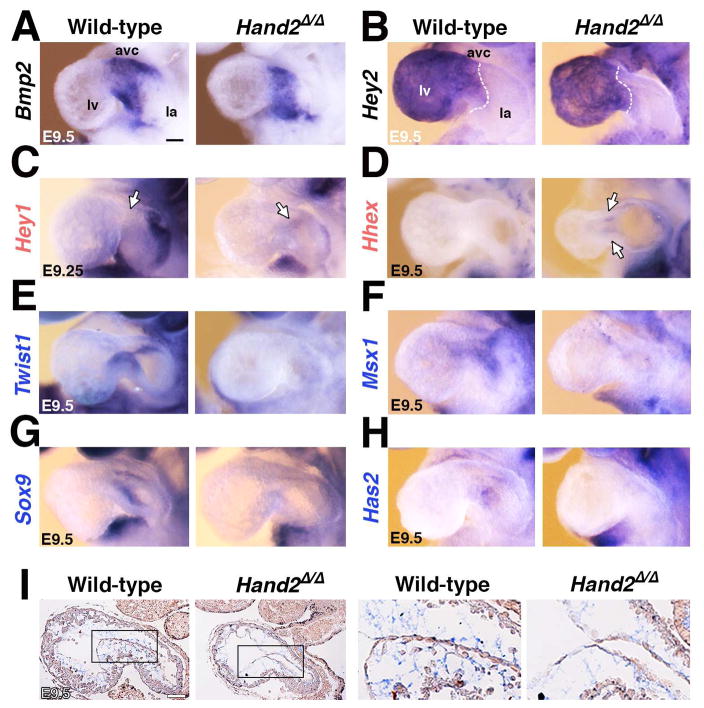
Figure 5. Whole mount in situ hybridization reveals spatial changes in some of the differentially expressed HAND2 target genes.
(A, B) Unaltered expression of Bmp2 (panel A) and Hey2 (panel B) indicates that the AVC domain is established correctly in Hand2-deficient hearts. (C, D) Ectopic expression of the Hey1 (panel C) and Hhex transcriptional regulators (panel D) in the mutant AVC corroborated their up-regulation detected by RNA-Seq analysis. (E-F) Loss of Twist1 (panel E), Msx1 (panel F), Sox9 (panel G) and Has2 (panel H) from the mutant AVC is in agreement with their transcriptional down-regulation detected by RNA-Seq analysis. (I) Alcian blue staining of glycosaminoglycans shows the reduced deposition of extra-cellular matrix in the cardiac jelly of Hand2-deficient mouse embryos. Right panels show the enlargements indicated by frames in the left panels. Gene names in black: unaltered, red: increased, blue: reduced transcript levels as determined by RNA-Seq analysis (Figure 4). Scale bars: 100μm. See also Figure S5.