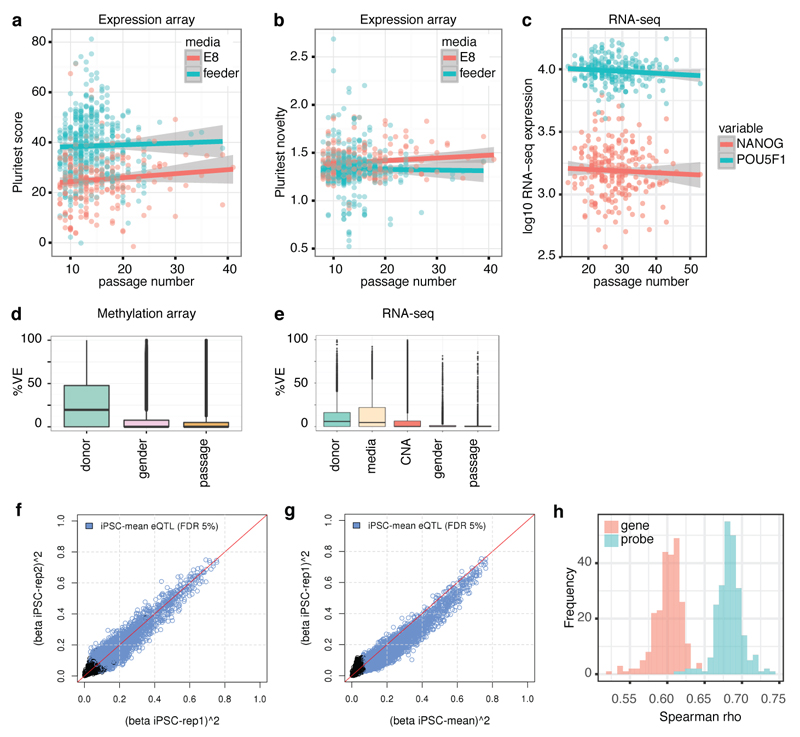
Extended Data Figure 6. Effect of passage on Tier 1 and Tier 2 data and overview of iPSC cis eQTLs mapped with ‘Tier 1’ gene expression array data.
(a,b) Passage number versus PluriTest pluripotency and novelty scores shows no significant association between passage number and pluripotency. Trend lines shown are fit using linear regression of PluriTest scores on passage number (score P = 0.66, novelty P = 0.21). Association was also not deemed significant when including gender and media as fixed effects and batch variables and donor as random effects (score P = 0.3, novelty P = 0.14). (c) Passage number versus log10 RNA-seq expression of pluripotency factors Nanog and Pou5f1 (Oct4) shows no significant association between passage number and pluripotency. Trend lines are fit using linear regression of log10 expression on passage number (Nanog P = 0.5, Pou5f1 P = 0.15). Association was also not deemed significant when considering the two genes together and when including gender and media as fixed effects and batch variables and donor as random effects (passage P = 0.28, passage-gene interaction P = 0.96). (d,e) Variance component analysis for Tier 2 assays, showing that for the majority of genes gender and passage explained little of the total variance. (f,g) Comparison of eQTL effect sizes (squared beta) at lead variants of the main gexarray eQTL map (derived using mean expression levels per donor). Plotted are the effect sizes for all tested genes (FDR < 5% eGenes indicated in blue) derived from (f) iPSC line replicate sets 1 and 2, one per donor, drawn randomly (rho = 0.47 genome-wide, rho = 0.80, FDR < 5% eGenes, P < 2.2e-16; Spearman rank correlation) and (g) replicate set 1 and the main map (rho = 0.57 genome-wide, rho = 0.88, FDR < 5% eGenes, P < 2.2e-16). Panel (g) shows that the effect sizes obtained using the mean expression values per donor are higher than when using individual lines. (h) Pairwise correlation between gene expression levels in iPSCs measured with RNA-seq and gexarray. Plotted are the Spearman rank correlation coefficients of either gene (pink) or gexarray probe (blue) region based read counts, demonstrating higher correlation of probe-based counts.