Abstract
Background & Aims
Transforming growth factor beta (TGFβ) acts either as a tumor suppressor or as an oncogene, depending on the cellular context and time of activation. TGFβ activates the canonical SMAD pathway through its interaction with the serine/threonine kinase type I and II heterotetrameric receptors. Previous studies investigating TGFβ-mediated signaling in the pancreas relied either on loss-of-function approaches or on ligand overexpression, and its effects on acinar cells have so far remained elusive.
Methods
We developed a transgenic mouse model allowing tamoxifen-inducible and Cre-mediated conditional activation of a constitutively active type I TGFβ receptor (TβRICA) in the pancreatic acinar compartment.
Results
We observed that TβRICA expression induced acinar-to-ductal metaplasia (ADM) reprogramming, eventually facilitating the onset of KRASG12D-induced pre-cancerous pancreatic intraepithelial neoplasia. This phenotype was characterized by the cellular activation of apoptosis and dedifferentiation, two hallmarks of ADM, whereas at the molecular level, we evidenced a modulation in the expression of transcription factors such as Hnf1β, Sox9, and Hes1.
Conclusions
We demonstrate that TGFβ pathway activation plays a crucial role in pancreatic tumor initiation through its capacity to induce ADM, providing a favorable environment for KRASG12D-dependent carcinogenesis. Such findings are highly relevant for the development of early detection markers and of potentially novel treatments for pancreatic cancer patients.
Keywords: Pancreas, Cancer, TGFβ, Acinar-to-Ductal Metaplasia, KRASG12D
Abbreviations used in this paper: ADM, acinar-to-ductal metaplasia; AFI, acinar fatty infiltration; EMT, epithelial-to-mesenchymal transition; PanIN, pancreatic intraepithelial neoplasia; PBS, phosphate-buffered saline; PDA, pancreatic ductal adenocarcinoma; RT-qPCR, reverse transcription quantitative polymerase chain reaction; TGFβ, transforming growth factor beta; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling
Graphical abstract
Summary.
Acinar-to-Ductal Metaplasia (ADM) is considered the main origin of pancreatic pre-neoplastic lesions that eventually develop into Pancreatic Ductal Adenocarcinoma (PDA). ADM could be a decisive step during tumorigenesis, selecting plastic cells for a more aggressive subsequent tumorigenesis. Our results indicate that TGFβ, a cytokine overexpressed during early and late pancreatic tumorigenesis, is an inducer of ADM and set up a favorable context for the emergence of oncogene-driven neoplastic lesions. This questions the usual dichotomist vision of TGFβ in PDA.
In mammals, the pancreas is a bifunctional organ.1 The exocrine pancreas, composed of acinar cells and ducts, accounts for more than 90% of the organ’s mass. Acinar cells secrete digestive enzymes that are further collected in a network of ducts that discharge pancreatic juices into the duodenum. The endocrine tissue, represented by islets of Langerhans embedded in the exocrine pancreas, controls blood glucose levels by secreting hormones such as insulin and glucagon. Embryonically, all pancreatic lineages arise from a multipotent progenitor present at day E9 in the mouse endoderm.2 The fate of the pancreas is dictated by the expression of progenitor markers such as SOX9 and CK19 for ductal cells, MIST1, PTF1A, CPA1, and ELA1 for acinar cells, and NGN3 for endocrine cells.2, 3, 4 Note that PTF1A is required for the specification of the pancreatic multipotent progenitor cells and is later expressed only in the adult acinar compartment.5
The adult pancreas is highly plastic6 to ensure organ integrity in response to internal (intracellular activation of digestive enzymes, obstruction of main ducts potentially as a result of gallstones) or external stresses (alcohol, trauma). Repair and regeneration of the injured organ are orchestrated by many cell types including acinar cells, centroacinar cells, ductal cells, immune cells, and stellate cells.7, 8 The existence of a resident stem cell population in the organ remains controversial.9, 10 Many studies1, 11, 12 have focused on post-injury pancreatic regeneration and demonstrated the crucial role of acinar cells during this process. Indeed, under severe stress conditions such as pancreatitis, acinar cells undergo acinar-to-ductal metaplasia (ADM), a morphologic and transcriptional conversion into duct-like cells with embryonic progenitor cell properties.5, 7, 13 These metaplastic cells are then capable of proliferating to replenish the damaged organ. In the case of a sustained stress signal or concomitant oncogenic activation such as KRAS activating mutations (eg, KRASG12D), the metaplastic cells cannot revert to a differentiated state, as generally observed in the case of an acute stress. This pancreatic erosion process constitutes a favorable setting for the onset of low-grade pancreatic intraepithelial neoplasia (PanIN).14 Progression toward PanINs of higher grade and eventually pancreatic ductal adenocarcinoma (PDA) is associated with recurrent mutations or genetic/epigenetic alterations in tumor suppressor genes (INK4/ARF, TP53).15, 16 Importantly, SMAD4/DPC4, a core component of the transforming growth factor beta (TGFβ) signaling pathway, is deleted in 50% of PDAs.16, 17, 18 Members of the TGFβ superfamily of transforming growth factors are involved in embryonic development, regulation of homeostasis, and the pathogenesis of a variety of diseases.19, 20 TGFβ signaling occurs through a heterotetrameric receptor complex composed of 2 subunits, the type I and type II TGFβ receptors (TβRI and TβRII, respectively). On binding to its receptors, TGFβ enables TβRII to transphosphorylate TβRI, which in turn activates the canonical SMAD pathway (phosphorylation of SMAD2 and SMAD3 that further interact with SMAD4 to accumulate inside the nucleus) and other signaling pathways (MAPK, RHOA, and PI3K/AKT).21 In cancer, TGFβ behaves as either a tumor suppressor or a tumor promoter. Indeed, TGFβ is generally considered to be a tumor suppressor early in tumor development (tumor initiation) by restricting epithelial cell growth (through cytostasis and apoptosis).19, 22, 23 However, in later stages of tumorigenesis (tumor progression), TGFβ has oncogenic properties through its capacity to regulate cellular plasticity by stimulating biological processes, including extracellular matrix deposition, immune evasion, epithelial-to-mesenchymal transition (EMT), and stemness.22, 24, 25, 26, 27
Unlike the majority of previously published studies in which the TGFβ signaling pathway was abrogated, our present work addresses the consequences of TGFβ cell-autonomous activation in the pancreatic epithelial lineage. To this end, we generated a unique mouse model enabling us to express a constitutively activated TβRI receptor (TβRICA) exclusively in pancreatic acinar cells, starting either in the embryo or after birth. Hence, we demonstrate that the specific activation of TGFβ signaling in pancreatic acinar cells induced ADM reprogramming and eventually facilitated the onset of KRASG12D-induced PanINs and PDA progression.
Materials and Methods
Mouse Strains and Handling
The LSL-TβRICA strain was previously generated in the laboratory of Dr Bartholin28, 29 (TβRICA is tagged with the hemagglutinin epitope at the C-terminus). We and others have functionally validated the TβRICA transgene after successful in vivo targeting in different subcellular compartments or organs such as immune cells,28, 30, 31, 32, 33, 34, 35 ovaries,36 and uterine tissue.37 In the present study, we only used TβRICA males to circumvent the mosaic phenotype occurring in females as a result of random X chromosome inactivation and associated inactivation of the transgene in a proportion of cells, as previously reported for other transgenes located on the X chromosome.38 LSL-KrasG12D,39 Pdx1-Cre,40 Ptf1a-CreERT2,5 and Rosa-CreERT241 alleles have been described previously. E2A-Cre42 allele has been described previously; [E2A-Cre+/+] or [E2A-Cre+/-] mice were crossed with [LSL-TβRICA] (R) mice.
E19.5 embryos were removed from [LSL-TβRICA] females (R) previously impregnated by [Pdx1-Cre] males (C). To this end, pregnant females (R; n = 11) were euthanized at 19.5 days post coitum, and embryos were collected (n = 92).
Five-week-old mice bearing the Ptf1a-CreERT2 allele along with LSL-TβRICA and/or LSL-KrasG12D transgenes were injected with tamoxifen (Sigma-Aldrich #T5648; St Louis, MO; 3 mg per injection) to induce recombination of the LSL-TβRICA and LSL-KrasG12D alleles. Animals from the 3-day cohort received 2 injections (day 1 and day 3) and were killed at day 4. Animals from the 3-week, 2-month, and 6-month cohorts received 5 injections (days 1, 3, 5, 7, and 9) and were killed 3 weeks, 2 months, and 6 months after the first injection, respectively. For Rosa26-CreERT2 mice, tamoxifen was injected 7-10 weeks after birth.
Mice were housed and bred in the AniCan specific pathogen-free animal facility of the Centre de Recherche en Cancérologie de Lyon, France. The experiments were performed in compliance with the animal welfare guidelines of the European Union and with French legislation (CECCAPP protocol #CLB-2014-008).
Histology and Immunohistochemistry/Immunofluorescence
Histologic (H&E staining) and immunohistochemical experiments were performed as described previously.43, 44 For immunohistochemistry experiments, the primary antibodies used were anti-CK19 (Developmental Studies Hybridoma Bank, Iowa City, IA) and anti-INSULIN (Dako A0564; Glostrup, Denmark). The secondary antibodies used were rat immunoglobulin G (H+L) biotinylated (Vector #BA-9400; Vector Laboratories, Burlingame, CA) and guinea pig immunoglobulin G (H+L) biotinylated (Vector #BA-7000). For AMYLASE/CK19 double immunofluorescence, primary antibodies were anti-AMYLASE (Sigma-Aldrich A8273) and anti-CK19 (Developmental Studies Hybridoma Bank) and secondary antibodies used were Rabbit IgG (H+L) Alexa Fluor 647-conjugated (Life Technologies #A-21245 GAR647) and Rat IgG (H+L) Alexa Fluor 594-conjugated (Life Technologies #A-21209 DAR594). For AMYLASE/SOX9 double immunofluorescence, primary antibodies were anti-AMYLASE (Santa-Cruz #166349), anti-SOX9 (Millipore #AB5535) and secondary antibodies used were Rabbit IgG (H+L) Alexa Fluor 647-conjugated (Life Technologies #A-21245 GAR647) and Mouse IgG (H+L) Alexa Fluor 488-conjugated (Life Technologies #A-11001 GAM488), but artificial colors (red for AMYLASE and green for SOX9) were given with the Zeiss software to be consistent with AMYLASE/CK19 double staining. Nuclei were counterstained with DAPI, and images were acquired with a Zeiss Imager M2 AX10 (Carl Zeiss AG, Oberkochen, Germany).
Quantification of Pancreatic Lesions
Histologic scoring of pancreatic lesions was performed by using 1 representative H&E tissue slide per animal (3–8 animals per condition). Pancreatic lesions were scored from PanIN1 to PanIN3/PDA and were counted. The area of the analyzed tissue was determined by using ImageJ software (National Institutes of Health, Bethesda, MD), and lesion counts were normalized to this area.
RNAscope
TβRICA mRNA and Smad7 mRNA were detected in situ by using the RNAscope technology (Advanced Cell Diagnostics, Newark, CA) for the Hu-TβRI (catalog no. 431041) and Mm-Smad7 (catalog no. 429411) probes, respectively.
Cell Culture
The rat pancreatic acinar cell line AR42J (ATCC) was cultured in Dulbecco modified Eagle medium supplemented with fetal calf serum (Lonza Group, Basel, Switzerland) and penicillin/streptomycin (Gibco Laboratories, Gaithersburg, MD). AR42J cells were infected with murine retroviral particles containing a wild-type (pZip-Neo) vector or a KRASG12D-expressing vector (pZIP-Neo-KRASG12D; donated by Dr J. Caramel, CRCL, Lyon, France) and further cultured in the presence of Geneticin (PAA Laboratories, Linz, Austria). TGFβ was used at the final concentration of 10 ng/mL.
For proliferation assays, AR42J-WT and AR42J–KRASG12D were seeded in triplicate at 100,000 cells per well onto 12-well plates and treated with TGFβ (10 ng/mL; PeproTech #100-21; Rocky Hill, NJ) for 24 and 48 hours. For each time point, cells were morphologically examined by phase-contrast microscopy and counted by using the trypan blue exclusion method. Kinase inhibitors were used as follows: 5 μmol/L TβRI inhibitor (SB 431542, Sigma-Aldrich #S4317), 10 μmol/L MEK inhibitor (U0126 monoethanolate, Sigma-Aldrich #U120), 2.5 μmol/L JNK inhibitor (Selleckchem #S4901; Houston, TX), 2.5 μmol/L PI3K inhibitor (LY294002, Selleckchem #S1105), and 2.5 μmol/L p38 inhibitor (SB 203580; Enzo Life Sciences, Farmingdale, NY; #BML-EI286-0001). Inhibitors were added 1 hour before TGFβ treatment (10 ng/mL) and maintained during the 48 hours of TGFβ treatment.
Immunoprecipitation
For protein analysis, mouse pancreas tissue was lysed and homogenized in RIPA buffer (50 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 1 mmol/L ethylenediamine tetraacetic acid, 0.5% sodium deoxycholate, 0.1% sodium dodecylsulfate, and 1% Nonidet) containing a cocktail of protease and phosphatase inhibitors. After protein quantification, 40 μg protein was used for loading controls, and 2.5 mg protein was used for the immunoprecipitation assay by using the SMAD2/3 antibody (Cell Signaling Technology #3102; Danvers, MA) and protein G Agarose Fast Flow (EMD Millipore). Proteins were separated by sodium dodecylsulfate–polyacrylamide gel electrophoresis and detected by Western blot analysis.
Western Blot Analysis
Western blotting was performed as previously described.29 The primary antibodies used were anti-phospho-SMAD2 (Cell Signaling #3101), anti-SMAD2 (Invitrogen #51-1300; Carlsbad, CA), anti-SMAD2/3 (Cell Signaling #3102), anti-SMAD4 (Epitomics #1676-1; Burlingame, CA), anti-RAS (Santa Cruz Biotechnology), anti-β-actin (Sigma-Aldrich #A5441 Clone AC15), and anti-β-tubulin (mouse monoclonal; Sigma-Aldrich). Peroxidase-labeled anti-rabbit and anti-mouse secondary antibodies were purchased from Dako (mouse immunoglobulin G horseradish peroxidase–conjugated DAKO P0260, rabbit immunoglobulin G horseradish peroxidase–conjugated DAKO #P0448).
Polymerase Chain Reaction and Reverse Transcription Quantitative Polymerase Chain Reaction
PCR to detect the recombined TβRICA allele was performed as described previously.28
AR42J-WT and AR42J-KRASG12D were treated for 48 hours with 10 ng/mL TGFβ (PreproTech #100-21) before being washed with phosphate-buffered saline (PBS). RNA extraction was performed by using the RNeasy mini kit (QIAGEN #74104; Hilden, Germany), according to the manufacturer’s instructions.
For pancreatic tissues, RNA extraction from frozen tissue was performed by using guanidine thiocyanate enriched lysis solution containing 5 mol/L guanidine thiocyanate (Sigma-Aldrich #G6639), 2.5 mmol/L sodium citrate, 0.5% N-lauryl sarcosine (Sigma-Aldrich #61739), and 1% β-mercaptoethanol. Lysates were centrifuged at 14,000 rpm at 4°C for 5 minutes to eliminate cell fragments. RNA was consecutively purified with the RNeasy mini kit (QIAGEN #74104), according to the manufacturer’s instructions.
After RT (ThermoFisher Scientific, Waltham, MA) by using random primers, qPCR was performed by using the MESA GREEN qPCR MasterMix Plus for SYBR Assay ROX (Eurogentec, Liège, Belgium; #RT-SY2X-06+WOU) and the following primers:
| Ela1: | Forward: 5′-TGGTCCTGTATGGACACAGT-3′ |
| Reverse: 5′-CCGTCATTCTGACTCAGGTT-3′ | |
| Mist1: | Forward: 5′- TGGCTAAAGCGACGTGTCC-3′ |
| Reverse: 5′- CTTCCGACTGGGGATCCGA-3′ | |
| Cpa1: | Forward: 5 ′- ACTTTGTGGGACACCAGGTT-3′ |
| Reverse: 5′- ACATCAATGGGGATACCGGC-3′ | |
| Sox9: | Forward: 5′- GTGCTGAAGGGCTACGACTGGA-3′ |
| Reverse: 5′- GTTGTGCAGATGCGGGTACTGG-3′ | |
| Hnf1β: | Forward: 5′- TCCCATCTGCAATGGTGGTC-3′ |
| Reverse: 5′- GCTGTGCACAAAGTGAGTGG-3′ | |
| Hes1: | Forward: 5′- GTCCCCGGTGGCTGCTAC-3′ |
| Reverse: 5′- AACACGCTCGGGTCTGTGCT-3′ | |
| Serpine-1: | Forward: 5′- CCCACGGAGATGGTTATAG -3′ |
| Reverse: 5′- ATCACTTGCCCATGAAGAG -3′ | |
| TβRICA: | Forward: 5′- TTGTGAACAGAAGTTAAGGC -3′ |
| Reverse: 5′- AGCATAATCAGGAACATCAT -3′ | |
| Actb: | Forward: 5′- GCAGGAGTACGATGAGTCCG-3′ |
| Reverse: 5′- ACGCAGCTCAGTAACAGTCC-3′ | |
| Bmf: | Forward: 5′- AGGTACAGATCGCCAGAAAGC-3′ |
| Reverse: 5′- CTCGGTTCTGCTGGTGTTGTTG-3′ |
RAS Activity Assay
GTP-bound RAS (activated RAS) was measured in cell lysates incubated with beads coated with RAF1-RBD (Upstate; EMD Millipore). The experiment was performed by using the active RAS detection kit (Cell Signaling Technology #8821).
Cell Death Assays
For the caspase-3 activity assay, AR42J-WT and AR42J-KRASG12D cells were treated with 10 ng/mL TGFβ for 12 hours. Caspase-3 activity was determined by using the CASPASE-3/CPP32 Fluorimetric Assay Kit, according to the manufacturer’s instructions (Gentaur Biovision, Kampenhout, Belgium). For the annexin V assay, AR42J-WT and AR42J-KRASG12D cells were seeded in duplicate and treated with 10 ng/mL TGFβ for 48 hours (PeproTech). Cells were harvested in PBS and incubated with annexin V–fluorescein isothiocyanate and propidium iodide (BD Pharmingen, San Diego, CA), according to the manufacturer’s instructions, and analyzed by flow cytometry (FACS Calibur cytometer; BD Biosciences). For the TUNEL cell death assay, AR42J-WT and AR42J-KRASG12D (24 hours of TGFβ treatment) cells or tissue sections were fixed in formaldehyde before being permeabilized with PBS-0.2% Triton X-100 and incubated in TdT buffer, 1 mmol/L CoCl2 (Sigma-Aldrich # 15862-1 mL-F) for 5 minutes. Then they were incubated with TdT buffer (30 mmol/L Tris, 150 mmol/L sodium cacodylate, pH7.5), 1 mmol/L CoCl2, Biotin16-dUTP (Sigma-Aldrich #11093070910), TdT enzyme (Roche #11767305001; Basel, Switzerland) for 1 hour at 37°C. The reaction was stopped by incubating the samples with 300 mmol/L NaCl, 2.5 mmol/L sodium citrate for 15 minutes. After washing with PBS, unspecific sites were saturated with 2% bovine serum albumin, 10 mmol/L PBS. After washing with PBS, slides were incubated with streptavidin-Cy3 (Jackson ImmunoResearch, West Grove, PA) diluted 1/200 in 2% bovine serum albumin in PBS. Finally, the nuclei were counterstained with DAPI (Sigma-Aldrich), and images were acquired with a Zeiss Imager M2 AX10.
Results
Transforming Growth Factor Beta-induced Cell Growth Inhibition In Vitro Is Enhanced in Acinar Cells Expressing KRASG12D
We initially explored the combined effects of KRAS activation and TGFβ treatment on rat AR42J pancreatic acinar cells45, 46 infected either with a wild-type retroviral vector (AR42J-WT cells) or with a KRASG12D retroviral vector (AR42J-KRASG12D cells). We ascertained that KRASG12D was present and functional in AR42J-KRASG12D cells via Western blot analysis, which revealed a significant increase in the level of active GTP-bound RAS protein compared with AR42J-WT cells (Figure 1A). Examination of the cells by phase-contrast microscopy revealed that AR42J-KRASG12D cells treated with TGFβ grew to a lower cell density than the other cell populations (Figure 1B). This growth-inhibition effect was confirmed by counting cells, highlighting a clear decrease in the number of AR42J-KRASG12D cells treated with TGFβ (Figure 1C). Furthermore, this decrease was much stronger than that observed for AR42J-WT cells. Functionally, although these latter cells were poorly responsive to TGFβ with respect to their capacity to phosphorylate SMAD2 (P-SMAD2), AR42J-KRASG12D cells displayed a drastic increase in P-SMAD2 on TGFβ treatment, as evidenced by Western blot analysis (Figure 1D). As expected, the levels of SMAD2/3 and SMAD4 were affected neither by KRAS activation nor by TGFβ treatment. Taken together, these results demonstrate that the activation of KRAS potentiates TGFβ cell growth inhibition and SMAD pathway activation in rat pancreatic acinar cells.
Figure 1.
KRASG12Dpotentiates TGFβ-mediated cell growth inhibition of AR42J rat pancreatic acinar cells. (A) Immunoblot of RAS-GTP and total RAS on AR42J-WT and AR42J-KRASG12D protein extracts. (B) Phase microscopy of AR42J-WT and AR42J-KRASG12D cells 48 hours after TGFβ treatment. Insets represent higher magnifications fields. Scale bar, 50 μm/L. NT, untreated. *Apoptotic cells. (C) Cell count of AR42J-WT and AR42J-KRASG12D cells at indicated times after TGFβ treatment. Representative experiment performed in duplicate (means ± standard deviation) is shown. (D) Immunoblot of SMAD proteins after 2-hour TGFβ treatment.
KRASG12D Sensitizes Acinar Cells to Transforming Growth Factor Beta–induced Apoptosis and Dedifferentiation In Vitro
Next, we speculated that the inhibition of AR42J-KRASG12D pancreatic acinar cell growth in the presence of TGFβ was due to apoptosis.47 Microscopic examinations revealed that TGFβ-treated AR42J-KRASG12D cells were characterized by the presence of poorly refringent cells (Figure 1B, inset) and by an increase in the number of floating cells (data not shown), evoking the presence of apoptotic cells. The highest rates of apoptosis were observed in AR42J-KRASG12D cells treated with TGFβ, as demonstrated by caspase-3 activity (Figure 2A), terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (Figure 2B), and annexin-V (Figure 2C) assays. TGFβ-induced apoptosis mainly occurs via the mitochondrial pathway by downregulating antiapoptotic factors of the BCL-2 family (BCL-xL) and by positively regulating proapoptotic factors of the BCL-2 family.23 Proapoptotic Bmf (BCL2-modifying factor) was previously shown to be upregulated after TGFβ treatment in the normal murine mammary epithelial cell line NMuMG.48 Reverse transcription quantitative polymerase chain reaction (RT-qPCR) experiments performed on total RNA prepared from AR42J-WT and AR42J-KRASG12D cells showed an increase of Bmf mRNA expression after TGFβ treatment (Figure 2D). This activation is compromised in the presence of SB431542, an inhibitor of TβRI kinase activity. These observations are consistent with the increase in apoptosis observed by caspase-3, TUNEL, and annexin-V assays.
Figure 2.
KRASG12Dand TGFβ cooperate to activate apoptosis of AR42J rat acinar cells. (A) Caspase-3 assay after 12-hour TGFβ treatment. (B) TUNEL assay after 24-hour TGFβ treatment. Upper panel, fluorescence microscopy. Lower panel, graphs showing percentage of TUNEL-positive cells under fluorescence microscopy and counted in each condition (>1000 cells). (C) Annexin-V/propidium iodide (PI) assay after 48-hour TGFβ treatment. Upper panel, raw FACS data. Upper right box drawn on each plot corresponds to apoptotic cell population (cells positive for both annexin-V and PI). Lower panel, quantification of apoptotic cells in upper right boxes drawn on plots above. For (A–C), representative experiment performed in triplicate (means ± standard deviation) is shown. (D) RT-qPCR of proapoptotic Bmf (BCL2-modifying factor) marker. Cells were treated (or not) with TGFβ ± TβRI inhibitor (SB431542) for 48 hours. For each condition, mRNA level is represented as mean ± standard deviation of 1 representative experiment performed in triplicate.
Phase-contrast micrographs revealed that TGFβ-treated AR42J-KRASG12D cells (and to a lesser extent AR42J-WT cells) acquired a spindle-like shape after TGFβ treatment (Figure 3A). RT-qPCR did not reveal significant changes in response to TGFβ in EMT markers such as Snai1, Zeb1, Vimentin, and Cdh2 in AR42J-WT and AR42J- KRASG12D cells (data not shown). In contrast, a decrease in the expression of acinar markers (Ela1, Cpa1, and Mist1) and a marked increase in the expression of ductal/dedifferentiation/progenitor markers (Hes1, Hnf1β, Sox9) were observed in AR42J- KRASG12D cells treated with TGFβ (Figure 3B). These morphologic and transcriptional changes are reminiscent of ADM. Among the genes we tested and known to be positively correlated to ADM, we focused our interest on Hnf1β because it was the best TGFβ responder in experiments presented in Figure 3B. TGFβ signaling is mediated by both the canonical SMAD pathway and non-canonical pathways (MAPK, PI3K, RHO-RAC).22, 23 AR42J-WT cells were cultured in the presence of TGFβ along with kinase inhibitors of both the canonical and non-canonical TGFβ pathways. RT-qPCR revealed that Hnf1β activation depends on SMAD and MEK pathways but not on JNK, P38, or PI3K pathways (Figure 3C).
Figure 3.
KRASG12Dand TGFβ cooperate to activate dedifferentiation of AR42J rat acinar cells. (A) High-magnification microscopy of rat acinar cells (AR42J). Phase-contrast microscopy of AR42J-WT and AR42J-KRASG12D cells treated or not with TGFβ for 48 hours. NT, untreated. Scale bars, 100 μm. (B) RT-qPCR of acinar (Ela-1, Cpa1, Mist1), progenitor (Hnf1β, Hes1), and ductal (Sox9) markers after 48-hour TGFβ treatment. For each condition, expression is represented as mean ± standard deviation of at least 3 independent experiments. Statistical analyses were performed by using Mann-Whitney test: *P < .05, **P < .01, ***P < .001. Non-significant (ns) if P > .05. (C) Signaling pathways involved in TGFβ-induced Hnf1β mRNA activation. AR42J-WT cells were cultured in presence of TGFβ along with different kinase inhibitors (inh) of canonical and non-canonical TGFβ pathways: TβRI inh, SB431542; MEK inh, U0126 monoethanolate; JNK inh, JNK-IN-8; P38 inh, SB203580; PI3K inh, LY294002. Hnf1β mRNA was detected by RT-qPCR after 48-hour treatment. For each condition, folds are represented as mean ± standard deviation of at least 3 independent experiments. Statistical analyses were performed by using Mann-Whitney U test: **P < .01. Nonsignificant (ns) if P > .05.
These results demonstrate that TGFβ treatment and KRAS activation cooperate to compromise AR42J rat acinar cell differentiation by increasing both apoptosis and ductal-like reprogramming, two hallmarks of ADM in vivo.
Activation of Transforming Growth Factor Beta Signaling in the Mouse Embryonic Pancreas Severely Compromises the Development of the Acinar Compartment
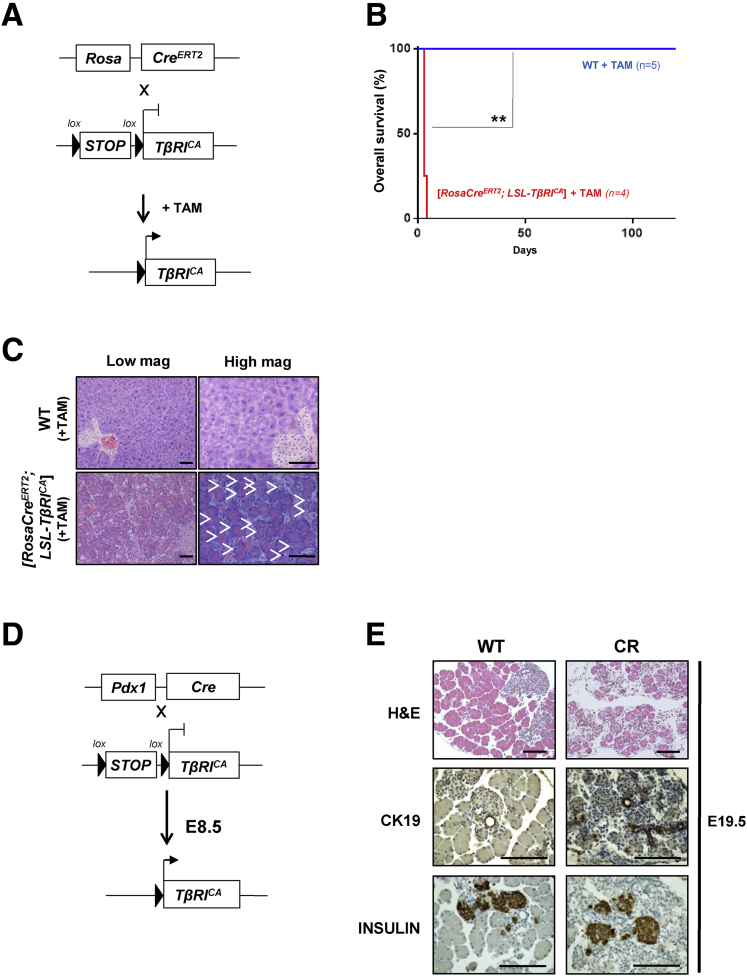
We then analyzed whether the activation of TGFβ signaling (alone or in combination with KRAS activation) could compromise the homeostasis of pancreatic acinar cells in vivo. To achieve this, we used a mouse strain ([LSL-TβRICA] also called R) containing a constitutively active TGFβ type I receptor (TβRI), previously generated in the laboratory of Dr Bartholin by using a knock-in strategy.28, 30 Transgene expression under the control of the ubiquitous CAG (human cytomegalovirus enhancer and chicken β-actin) promoter is repressed by a floxed transcriptional Stop (LSL, Lox-Stop-Lox), which can be excised in the presence of Cre recombinase. E2A-Cre allows recombination early during embryonic development before the E5 uterine wall implantation stage. We never obtained [E2A-Cre; LSL-TβRICA] mice (Figure 4), strongly suggesting that the expression of TβRICA at an early stage of development was embryonically or prenatally lethal. To restrict TGFβ gain-of-function to the whole organism of mice postpartum, we generated [Rosa-CreERT2; LSL-TβRICA] mice that were subsequently treated with tamoxifen to induce ubiquitous Cre-mediated recombination, the Rosa-26 locus being active in all adult lineages (Figure 5A). All [Rosa-CreERT2; LSL-TβRICA] mice (n = 4) injected with tamoxifen at the age of 7–10 weeks were euthanized 4 or 5 days after treatment because of the rapid onset of a highly detrimental phenotype, whereas all of the wild-type mice survived (n = 5) (Figure 5B). Histologic examination of the internal organs revealed that the pancreas was the most impaired organ, with many apoptotic figures (Figure 5C), clearly indicating a deleterious effect of TβRI activation in the pancreas, which could explain the severe phenotype of these animals. To test this hypothesis, we expressed TβRICA in all of the pancreatic epithelial lineages by crossing [LSL-TβRICA] (R) and [Pdx1-Cre] (C) mice,40 which drives Cre recombinase expression in embryonic progenitors (at E8.5) of all pancreatic epithelial lineages (Figure 5D). Histologic analysis revealed that [Pdx1-Cre; LSL-TβRICA] embryos (CR) presented pancreatic defects characterized by a massive reduction in the number of mature acinar cells (hematoxylin-eosin staining) along with abnormally abundant ductal structures (shown by CK19 expression) (Figure 5E). No obvious defect in the endocrine compartment was observed (using INSULIN staining as a marker) (Figure 5E). These latter results show that pancreatic activation of TGFβ signaling at an early stage of development affects normal pancreatic development by impacting the acinar cell compartment.
Figure 4.
Early activation of TβRICAduring mouse development is embryonically lethal. [E2A-Cre+/+] or [E2A-Cre+/-] mice were crossed with [LSL-TβRICA] (R) mice. Total number of litters, pups, and offspring genotype distribution are presented. Fisher’s exact test was performed to statistically confirm absence of [E2A-Cre+/-; LSL-TβRICA] mice. Nonsignificant (ns), P > .05; *P < .05.
Figure 5.
Expression of TβRICAin mouse embryo compromises development of acinar compartment. (A) Breeding strategy to target TβRICA expression in whole adult body by using tamoxifen-inducible Rosa26-CreERT2 allele. (B) Overall survival (Kaplan-Meier analysis) of wild-type (WT) and [Rosa26-CreERT2; LSL-TβRICA] mice after tamoxifen injection. Log-rank (Mantel-Cox) test. **P = .0047. (C) Histology of pancreata prepared from WT and [Rosa26-CreERT2; LSL-TβRICA] mice 5 days after tamoxifen injection. White arrows, apoptotic cells. Scale bars, 200 μm. (D) Breeding strategy to target TβRICA expression in all epithelial pancreatic lineages from embryonic day 8.5 (E8.5) by using Pdx1-Cre allele. (E) Histology of pancreata prepared from WT and [Pdx1-Cre; LSL-TβRICA] (CR) E19.5 embryos. Scale bars, 200 μm. mag, magnification; TAM, tamoxifen.
Inducible TβRICA Expression in Pancreatic Acinar Cells After Birth Results in SMAD Pathway Activation
To circumvent the afore-mentioned embryonic developmental defects, we investigated the effects of TβRICA expression starting after birth. To that end, we used the tamoxifen-inducible Pft1a-CreERT2 mouse allele (CER),5 which limits the expression of Cre recombinase to adult acinar cells (Figure 6A). Mice were injected with tamoxifen at 5 weeks of age. Excision of the floxed transcriptional Stop in the pancreas of CERR(+TAM) mice was validated by PCR on genomic DNA (Figure 6B). RT-qPCR experiments revealed a significant enrichment in TβRICA mRNA in CERR(+TAM) pancreata compared with R pancreata, which was correlated with upregulation of the TGFβ-target gene Serpine-1 (Figure 6C). Immunoprecipitation assays and Western blot analyses revealed a marked accumulation of P-SMAD2 in the pancreata of CERR(+TAM) compared with wild-type mice (Figure 6D) (as expected the level of total SMAD2 in the lysates or in the immunoprecipitates did not differ between wild-type and CERR(+TAM) mice). Next, we ascertained that the expression of the TβRICA transgene was restricted to the pancreatic acinar compartment by performing RNAscope (Figure 6E). It is worth noting that 100% of the acinar cells expressed the transgene, and that transgene expression was not observed in other lineages (ducts and islets). In addition, the detection of Smad7 mRNA by RNAscope suggested that the canonical TGFβ-SMAD pathway was activated in vivo in the presence of TβRICA. These results clearly indicate that the conditional TβRICA transgene is restricted to pancreatic epithelial cells on tamoxifen treatment and is correlated with the activation of the SMAD pathway in vivo.
Figure 6.
Inducible TβRICAexpression in pancreatic acinar cells after birth results in SMAD pathway activation in vivo. (A) Breeding strategy to express TβRICA after birth in pancreatic acinar cells by using tamoxifen-inducible Pft1a-CreERT2 allele. Black arrows represent genotyping primers. Sizes of expected fragments are shown. (B) PCR on genomic DNA to assess excision of STOP signal in CERR pancreas after tamoxifen injection. (C) RT-qPCR of TβRICA and Serpine-1 on total RNA from pancreata prepared from mice treated with tamoxifen. Expression level in R mice was arbitrarily set at 1 (mean ± standard deviation; 2 independent experiments). (D) Immunoprecipitation of SMAD2/3 and Western blot analysis of P-SMAD2 (phospho-SMAD2) and total SMAD2. Total lysate was assessed for β-tubulin and SMAD2. (E) RNAscope detection of TβRICA and Smad7 mRNA on pancreatic sections from WT and CERR mice. Scale bars, 100 μm. (A–E): WT, wild-type; R, [LSL-TβRICA]; CERR, [Pft1a-CreERT2; LSL-TβRICA]. mag, magnification; TAM, tamoxifen.
Acinar-to-Ductal Metaplasia in Response to TβRICA Expression
Activating mutations of KRAS (eg, KRASG12D) are found in >90% of human PDAs and are sufficient to trigger carcinogenesis when targeted in mice.49 We generated mice expressing TβRICA (R) and KrasG12D (K) transgenes either alone or in combination under the control of Pft1a-CreERT2 (CER), and we performed histologic analyses of pancreata collected at different time points after tamoxifen injection (3 days, 3 weeks, 2 months, and 6 months). As early as 3 days after tamoxifen treatment (Figure 7A), CERR(+TAM) and CERKR(+TAM) pancreata presented a severe reduction in the acinar compartment associated with increased apoptosis (Figure 7B; H&E, black arrowheads; TUNEL). CERK(+TAM) were normal and CERKR(+TAM) undistinguishable from CERR(+TAM) pancreata. Amylase is a marker of acinar cells, whereas CK19 and SOX9 are ductal markers. SOX9 is also an early positive regulator of ADM. Interestingly, we observed that CERR(+TAM) and CERKR(+TAM) pancreata displayed double-positive CK19+/amylase+ or SOX9+/amylase+ acinar cells (Figure 7B, immunofluorescence, white arrowheads), a feature of acinar cells undergoing early ADM reprogramming to form ductal-like structures.50 These “ducts” consisted of simple cuboidal epithelial cells and were characterized by the expression of stemness markers and ductal markers. These double-positive acinar cells (CK19+/amylase+, SOX9+/amylase+) most likely correspond to a regenerative process accompanying acinar cell loss. Three weeks after tamoxifen treatment (Figure 8), CERK(+TAM) pancreata were still normal. However, compared with the analysis performed 3 days after tamoxifen injection, we observed a drastic loss in the acinar compartment (amylase+), concomitantly with a large increase in the number of duct-like structures (CK19+), when TβRICA was expressed (CERR(+TAM)), independently of oncogenic KRAS activation (eg, KRASG12D).
Figure 7.
TβRICAexpression in acinar cells induces apoptosis and ductal-like differentiation 3 days after induction. (A) Diagram of experimental design for 5-week-old mice injected with tamoxifen and euthanized 3 days later. (B) H&E staining, TUNEL assay, and immunofluorescence of amylase, CK19, and SOX9. Black arrowheads, apoptotic cells; white arrowheads, CK19/amylase and SOX9/amylase double-positive cells; WT, wild-type; [Pft1a-CreERT-; LSL-TβRICA], CERR; [Pft1a-CreERT2; LSL-KrasG12D], CERK; [Pft1a-CreERT2; LSL-TβRICA; LSL-KrasG12D], CERKR. TAM, tamoxifen. Scale bars, 50 μm.
Figure 8.
TβRICAexpression in acinar cells leads to regenerative ADM 3 weeks after induction. (A) Diagram of experimental design for 5-week-old mice injected with tamoxifen and euthanized 3 weeks later. (B) H&E staining and immunofluorescence of amylase and CK19. WT, wild-type; [Pft1a-CreERT-; LSL-TβRICA], CERR; [Pft1a-CreERT2; LSL-KrasG12D], CERK; [Pft1a-CreERT2; LSL-TβRICA; LSL-KrasG12D], CERKR. TAM, tamoxifen. Scale bars, 100 μm.
Tumorigenesis Induced by KRASG12D Is Enhanced by TβRICA
Two and 6 months after tamoxifen treatment (Figure 9A and B), we observed in CERR(+TAM) pancreata a massive loss of acinar tissue replaced by adipose tissue, a phenomenon known as acinar fatty infiltration (AFI). In CERK(+TAM) pancreata, PanINs and ADM lesions were clearly distinguishable without clear evidence of AFI. Interestingly, CERKR(+TAM) double mutant pancreata presented both AFI and PanINs. Precise quantitative and qualitative analysis of epithelial lesions (Figure 9C) revealed at 2 months a 3-fold increase in the number of PanIN1 in CERKR(+TAM) compared with CERK(+TAM) and the absence of PanIN2 in CERK(+TAM), whereas these lesions were detectable in CERKR(+TAM). At 6 months the analysis showed the presence of PanIN3/PDA only in CERKR(+TAM) (absence in CERK(+TAM)), with three-fourths of CERKR(+TAM) mice developing PDA compared with 0 of 5 CERK(+TAM) mice (Fisher’s exact test, P = .0476; data not shown).
Figure 9.
TβRICAexpression in acinar cells accelerates KRASG12D-induced tumorigenesis several months after induction. (A) Diagram of experimental design representing 5-week-old mice injected with tamoxifen and euthanized 2 or 6 months later. (B) H&E staining. WT, wild-type; [Pft1a-CreERT-; LSL-TβRICA], CERR; [Pft1a-CreERT2; LSL-KrasG12D], CERK; [Pft1a-CreERT2; LSL-TβRICA; LSL-KrasG12D], CERKR. Scale bars, 100 μm. (C) Quantification of pancreatic epithelial lesions at different grades and observed in CERK and CERKR pancreata 2 months and 6 months after TAM injection. TAM, tamoxifen.
In double mutants, a strong desmoplastic reaction associated with a drastic reduction of the acinar compartment was observed at 2 and 6 months. Immunohistochemistry analysis of pancreata at 2 months and 6 months after tamoxifen induction confirmed the ductal nature of the lesions and demonstrated that TβRICA expression in CERR(+TAM) mice does not further affect endocrine islets compared with CERK(+TAM), which we have previously shown to present disorganized islets architecture51 (Figure 10).
Figure 10.
KRASG12Dand TGFβ activation cooperate to accelerate pancreatic tumorigenesis without affecting endocrine compartment. Immunohistochemical detection of CK19 and INSULIN in mice of indicated genotypes 2 and 6 months after tamoxifen induction. [Pft1a-CreERT2; LSL-TβRICA] (CERR); [Pft1a-CreERT2; LSL-KrasG12D] (CERK); [Pft1a-CreERT2; LSL-TβRICA; LSL-KrasG12D] (CERKR). Scale bars, 200 μm.
Our results demonstrate that in adult mice (1) activation of TGFβ signaling induces ADM by disrupting acinar cell homeostasis, leading to a drastic increase in the number of ductal structures at the expense of acinar structures; and (2) activation of TGFβ signaling in combination with oncogenic KRAS activation leads to the early onset of pre-neoplastic lesions that can naturally evolve toward high-grade/locally invasive lesions.
Discussion
Tumorigenesis can be divided into 3 consecutive steps. The priming stage corresponds to biological processes (such as cellular stress, inflammation, dedifferentiation) that predispose normal cells to further transformation by creating a propitious cellular state or microenvironment. The initiation stage is represented by all genetic and epigenetic events sufficient for the acquisition of the transformed phenotype (such as KRAS activating mutations). The progression/metastatic stage is characterized by mechanisms conferring aggressiveness and invasive properties to cancer cells (such as inactivation of TP53 or SMAD4 tumor suppressors). Bioactive TGFβ, which is found in normal and pathologic pancreas tissues, plays a crucial role in both normal tissue homeostasis and pancreatic diseases. The TGFβ paradox in cancer consists in the observation that TGFβ behaves either as a tumor suppressor or as a tumor promoter depending on the cellular context, especially in the 3 above-described stages. The studies conducted so far to address the multifaceted role of TGFβ in pancreatic cancer in vivo mostly relied on loss-of-function approaches (receptor inactivation, SMAD inactivation) and ligand gain-of-function (effect is not restricted to cancer cells). These strategies did not provide a clear answer as to the precise role of TGFβ signaling in pancreatic epithelial cells. In the present study, we developed a conditional and inducible TGFβ gain-of-function mouse model expressing a constitutively active type I TGFβ receptor (TβRICA) in the pancreas. We demonstrated that cell-autonomous expression of TβRICA in the epithelial pancreatic compartment could severely compromise acinar cell homeostasis by inducing early ADM reprogramming. This phenotype was associated with the activation of both a proapoptotic program and a ductal-like differentiation program. Lately, the predominant role of necroptosis (a programmed form of necrosis death) in acinar cell death was reported in severe experimental mouse pancreatitis.52 We also demonstrated that in the presence of mutated KRASG12D, TGFβ-induced ADM reprogramming facilitates the onset of PanINs. This work represents a demonstration that the activation of cell-autonomous TGFβ signaling compromises pancreatic acinar cell identity and eventually potentiates KRASG12D-driven tumor initiation.
Cell-autonomous Transforming Growth Factor Beta Activation Induces Acinar-to-Ductal Metaplasia Reprogramming
Three days after TβRICA induction in adult mouse pancreatic acinar cells, we observed a drastic reduction in acinar tissue as attested by decreased amylase expression in the pancreata of mice expressing the transgene. This observation corroborates previous studies, which reported a repressive role for TGFβ on the fate of acinar cells. Indeed, it was demonstrated that TGFβ could inhibit the formation of acinar tissue ex vivo in cultures of pancreatic embryonic buds.53 In vivo, by using a transgenic mouse model expressing a type II TGFβ dominant-negative mutant receptor under the regulation of the metallothionein 1 (Mt1) promoter, TGFβ was shown to be essential for the maintenance of the acinar compartment homeostasis.54 In the present study, we observed that targeted activation of TGFβ signaling in acinar cells in vivo efficiently compromised acinar identity. At the microscopic level, we observed that the loss of acinar tissue was associated with massive apoptosis and with the appearance of new highly abundant ductal structures of small diameter. This phenotype is reminiscent of ADM, a process involved in pancreas replenishment after tissue injury.1, 6, 11 During this process and as observed in the model presented here, the acinar cells are reprogrammed into a ductal-like cell population, displaying features of progenitor cells, with the ability to regenerate the different lineages in the injured pancreas. Recently, Liu et al55 have shown that TGFβ could convert primary human acinar cells to ductal-like cells in vitro, which corroborates our current in vivo demonstration that TGFβ induces ADM. Although the signals triggering ADM in vivo are poorly known, we demonstrate herein that TGFβ signaling activation is one of these signals.
Acinar-to-Ductal Metaplasia Reprogramming Induced by Transforming Growth Factor Beta Activation Facilitates KRASG12D-mediated Carcinogenesis
ADM has largely been documented to represent the first step in pancreatic tumor development by providing a suitable surrounding for the malignant transformation of cells in response to oncogene-driven mutations such as KRASG12D.12 Here we report a significant increase in both the number and the grade of PanIN lesions in the pancreas of mice expressing both TβRICA and KrasG12D transgenes in comparison with age-matched KRASG12D mice. PanINs were never observed in the presence of TβRICA alone. This observation supports the idea that KRASG12D generates a persistent ADM (known as for acinar-to-ductal reprogramming56), facilitating the subsequent onset of PanINs. Active TGFβ thus appears to be crucial to potentiate KRASG12D transforming properties through its capacity to confer a ductal-like phenotype to acinar cells that possess progenitor properties and that are more sensitive to transformation. It is worth underlining that the phenotype we observed in the pancreas of TβRICA mice resembles that of pancreatitis, a pathology predisposing to PDA.57 In aging mice we observed that ADM was resolved by AFI, which represents the normal evolution of chronic pancreatitis when the inflammatory stress ceases. Pancreatitis characterized by ADM has also been reported to be tightly dependent on TGFβ activation. Indeed, it was shown by others that inhibition of TGFβ in different mouse models, by using a dominant-negative type II TGFβ receptor,58, 59, 60 by overexpressing the inhibitory SMAD7,61 or by using halofuginone to inhibit TGFβ-induced collagen deposition,62 could compromise cerulein-induced pancreatitis. Pancreatitis has also been shown to be reversible in the absence of activated KRAS and irreversible in the presence of activated KRAS,63 demonstrating that KRASG12D is able to harness the pancreatitis phenotype to facilitate the development of PanINs. The presence of activated KRAS would then prevent spontaneous resorption of the phenotype, driving cells toward persistent ADM and PanINs. Our findings, together with these previous studies, demonstrate that TGFβ activation in the pancreas induces ADM, providing a propitious environment, with features reminiscent of pancreatitis, for the onset of KRASG12D-induced PanINs. Hence, cell-autonomous activation of TGFβ signaling in pancreatic acinar cells may represent a crucial step in pancreatitis, which is also known to predispose to PDA, thus providing a physiopathologic relevance for the results described in the present study.
Simultaneous Transforming Growth Factor Beta Signaling and KRAS Activation Cooperate to Induce Apoptosis and Ductal Reprogramming of Acinar Cells
We demonstrated in vitro and in vivo that the simultaneous activation of TGFβ and KRAS could cooperate to induce apoptosis and ductal reprogramming of acinar cells, two cellular events classically observed in pancreatic ADM.7 Importantly, in the tamoxifen-inducible model, 100% of pancreatic epithelial cells expressed the TβRICA transgene in vivo, thus indicating that the deleterious phenotype observed in the presence of the KRASG12D transgene resulted from a cell-autonomous functional interaction. Although our observations indicate that TGFβ activation induces direct ADM reprogramming, the mechanisms dictating cell fate, ie, apoptotic death versus survival associated with the acquisition of an ADM phenotype, remain to be uncovered. Indeed, the mechanisms that allow a proportion of acinar cells to escape apoptosis and pursue their route toward ADM remain to be identified. As suggested by a recent study, the fate of pancreatic cancer cells is tightly controlled by factors downstream of TGFβ (SNAIL and SOX4).64 David et al64 showed that TGFβ-induced EMT resulted in apoptosis activation and tumor suppressive activity. Hence, in the transformed pancreas, TGFβ-induced apoptosis reduces the number of cancer cells, thus substantiating its tumor suppressive effect at this stage, whereas in the normal pancreas (the present work), TGFβ induced apoptosis and promotes ADM, thereby proving its oncogenic effect. Whether this model proposed by David et al can be transposed to untransformed cells to study tumor initiation needs to be tested.
On the basis of the use of AR42J rat acinar cells stably expressing KRASG12D and treated with TGFβ, we were able to show that KRAS activation sensitized acinar cells to both TGFβ-induced apoptosis and dedifferentiation. At the molecular level, we evidenced in this model a cooperative effect of both pathways to modulate the expression of Hnf1β, Sox9, and Hes1. The combination of these 3 transcription factors is a hallmark of the formation of progenitor cells during development,3 which is in accordance with the well-documented role of TGFβ as a promoter of stemness.65, 66 Indeed, SOX9 and HNF1β are the first transcription factors that commit endoderm cells to pancreatic progenitors during organogenesis.5 They are markers of the ductal tree, re-expressed during ADM.3, 14 Moreover, the adult SOX9 has been shown to be a master positive regulator of the ADM process.56 Indeed, the ectopic expression of SOX9 in acinar cells activates the expression of ductal markers and increases KRASG12D-induced ADM, and conversely, inactivation of Sox9 in KRASG12D-expressing adult acinar cells compromises the onset of PanINs, confirming its critical role in the transition toward a ductal phenotype.56 In the present study, we reported that the in vivo co-expression of TβRICA and KRASG12D was able to induce the onset of amylase/SOX9 double-positive cells. This feature is known to be associated with the regenerative process after acinar cell loss, stemming from double-positive cells for acinar and ductal markers. The precise role of SOX9 in this model remains to be explored and whether Hes1 and Hnf1β are direct transcriptional targets of SMAD proteins. How KRASG12D potentiates their activation is currently under investigation in our laboratory.
Physiopathologic Relevance and Therapeutic Implications
Our observation that TGFβ behaves as a tumor promoter may seem paradoxical in light of other studies showing that inactivation of TGFβ signaling by inactivating Smad4 or TβRII accelerates the progression of KRASG12D-induced lesions.67, 68, 69, 70, 71, 72 This is most likely a consequence of both the gain-of-function approach we used and the stage at which TGFβ signaling was impaired. Indeed, in our model through its capacity to induce ADM, activation of TGFβ signaling resulted in an increased KRASG12D-induced tumor initiation. This can be explained by the effect of TGFβ on the priming stage of tumor progression by committing acinar cells to an ADM differentiation program favorable to transformation. In contrast, TβRII or Smad4 homozygous deletions facilitate the progression of KRASG12D-induced preexisting PanINs. This observation is consistent with the loss of SMAD4 at late stages and widespread metastatic human diseases.73, 74 Furthermore, we recently demonstrated that the activation of TGFβ signaling was sufficient to induce the onset of ovarian tumors, strongly supporting our current findings that this signaling pathway plays an active role in promoting tumor initiation.36 Overall, this work sheds new light on another level of complexity for the dual role of TGFβ as a tumor promoter and suppressor.
TGFβ or components of its signaling pathway are being extensively evaluated as a potential therapeutic target, as attested by the compelling preclinical and encouraging clinical studies.75 As a consequence of its dual role in cancer, deciphering TGFβ-related functional processes is a prerequisite to develop efficient anti-TGFβ therapies. Understanding the first steps of pancreatic tumorigenesis is necessary to determine how pancreatic tumor cells acquire plasticity. Such an effort may provide new therapeutic strategies aimed at restoring a normal differentiated state. An example of a successful differentiation therapy was observed in acute promyelocytic leukemia,76, 77 characterized by the accumulation of incompletely differentiated leukemic cells, which could be forced to fully differentiate on treatment with all-transretinoic acid. More recently, Fitamant et al78 provided proof-of-concept for differentiation therapy in solid tumors by deleting Yap1 in hepatocellular carcinoma, which present features of progenitor cells. In this context, unveiling the specific effects of TGFβ at different stages of tumorigenesis may lead to innovative therapeutic strategies aimed at restoring acinar differentiation and, therefore, at decreasing the deleterious onset of cellular plasticity. Defining the molecular mechanisms underlying the initiation of pancreatic cancer is highly relevant for the development of early detection markers and new therapies. Indeed, inhibition of TGFβ may represent a therapeutic strategy for impeding ductal reprogramming of acinar cells to prevent the initiation of PDA in high-risk patients (hereditary syndrome or chronic pancreatitis).
In conclusion, the present study demonstrates that TGFβ is an ADM inducer, facilitating the development of pancreatic neoplastic lesions in a KRASG12D-dependent context. According to the activated state of KRAS and TGFβ signaling, we propose an integrated and dynamic model for pancreatic cancer initiation; TGFβ activation is not sufficient to induce PanINs, and KRASG12D is poorly efficient at inducing ADM. However, when combined, TGFβ induces ADM and provides a propitious setting for KRASG12D-induced transformation (Figure 11). Defining the molecular mechanisms underlying the initiation of pancreatic cancer is highly relevant for the development of early detection markers and of potentially novel treatments.
Figure 11.
Schematic diagram showing effect of TGFβ signaling activation (alone or in combination with KRASG12D) on pancreatic acinar compartment. A few days after tamoxifen induction, TβRICA induces acinar cell apoptosis and subsequent regenerative metaplasia (ADM). In aging mice, ADM is overcome and replaced by AFI. KRASG12D activation alone induces ADM and PanINs several months after induction. When TβRICA and KRASG12D are simultaneously induced, PanINs develop much earlier. In the context of TβRICA-induced ADM, KRASG12D induces regenerative cells to undergo the ADM>ADR>PanIN sequence (ADR, acinar-to-ductal reprogramming). Hence, TGFβ signaling activation enhances the properties of KRASG12D through its capacity to prime a suitable surrounding for transformation. TAM, tamoxifen. [Pft1a-CreERT-; LSL-TβRICA], CERR; [Pft1a-CreERT2; LSL-KrasG12D], CERK; [Pft1a-CreERT2; LSL-TβRICA; LSL-KrasG12D], CERKR. TAM, tamoxifen. WT, wild-type.
Acknowledgments
The authors thank Dr Christophe Vanbelle for expert analyses in microscopy; the specific pathogen-free animal facility platform AniCan, ALECs-SPF facility and the Laboratoire des Modèles Tumoraux platform for animal care; the research and scientific services at the CRUK Beatson Institute in general; the laboratory of Dr Hingorani at the Fred Hutchinson Cancer Research Center for the help they provided during the revisions; and Christopher Wright for sharing the Ptf1a-CreERT2 mice with us.
Footnotes
Author Contributions Study concept and design were performed by NC, DFV, RMP, LBA, and LB. Acquisition of data was performed by NC, DFV, RMP, LBA, Jo.G, KY, CC, VC, ER, BK, SM, and CC. Analysis and interpretation of data were performed by NC, DFV, RMP, LBA, IT, VR, AC, OJS, UV, SS, PD, and LB. Drafting of the manuscript was performed by NC, DFV, and LB. Critical revision of the manuscript for important intellectual content was performed by NC, DFV, RMP, VR, AC, JM, Ju.G, OJS, UV, SS, PD, and LB. SS, LB and OJS obtained funding. Technical support was performed by SGL, AC, JV, NG, CC, JV, ES, and IG. LB supervised the study.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Association pour la Recherche sur le Cancer (JG, DFV, and BK salaries), the Centre Léon Bérard (SM and CK salaries), the Ministère de l’Enseignement Supérieur et de la Recherche of France (DFV, RMP, and ER fellowships), by the Ligue Nationale Contre le Cancer (VC, LA, and LBA fellowships), from the Ecole Normale Supérieure of Lyon (NC fellowship), and by Stem Cells Arabia (KY salary). LB is supported by an Avenir grant (Inserm), an INCA grant, Ligue grants (Rhône, Haute-Savoie), and an ARC grant. SS is supported by a Ligue grant (Ardèche). OJS is supported by a Cancer Research UK core grant (A21139) and an ERC Starting grant (311301). DFV is supported by an ERC Starting grant (311301).
References
- 1.Puri S., Folias A.E., Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell. 2015;16:18–31. doi: 10.1016/j.stem.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen M.C., Ahnfelt-Ronne J., Hald J., Madsen O.D., Serup P., Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 3.Shih H.P., Wang A., Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol. 2013;29:81–105. doi: 10.1146/annurev-cellbio-101512-122405. [DOI] [PubMed] [Google Scholar]
- 4.Pan F.C., Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 5.Pan F.C., Bankaitis E.D., Boyer D., Xu X., van de Casteele M., Magnuson M.A., Heimberg H., Wright C.V. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140:751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziv O., Glaser B., Dor Y. The plastic pancreas. Dev Cell. 2013;26:3–7. doi: 10.1016/j.devcel.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Murtaugh L.C., Keefe M.D. Regeneration and repair of the exocrine pancreas. Annu Rev Physiol. 2015;77:229–249. doi: 10.1146/annurev-physiol-021014-071727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beer R.L., Parsons M.J., Rovira M. Centroacinar cells: at the center of pancreas regeneration. Dev Biol. 2016;413:8–15. doi: 10.1016/j.ydbio.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey J.M., DelGiorno K.E., Crawford H.C. The secret origins and surprising fates of pancreas tumors. Carcinogenesis. 2014;35:1436–1440. doi: 10.1093/carcin/bgu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel E.V., Simeone D.M. Biology and clinical applications of pancreatic cancer stem cells. Gastroenterology. 2013;144:1241–1248. doi: 10.1053/j.gastro.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 11.Stanger B.Z., Hebrok M. Control of cell identity in pancreas development and regeneration. Gastroenterology. 2013;144:1170–1179. doi: 10.1053/j.gastro.2013.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills J.C., Sansom O.J. Reserve stem cells: differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci Signal. 2015;8:re8. doi: 10.1126/scisignal.aaa7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen J.N., Cameron E., Garay M.V., Starkey T.W., Gianani R., Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Roy N., Hebrok M. Regulation of cellular identity in cancer. Dev Cell. 2015;35:674–684. doi: 10.1016/j.devcel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macgregor-Das A.M., Iacobuzio-Donahue C.A. Molecular pathways in pancreatic carcinogenesis. J Surg Oncol. 2013;107:8–14. doi: 10.1002/jso.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan D.P., Hong T.S., Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 17.Hahn S.A., Schutte M., Hoque A.T., Moskaluk C.A., da Costa L.T., Rozenblum E., Weinstein C.L., Fischer A., Yeo C.J., Hruban R.H., Kern S.E. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 18.Knudsen E.S., O'Reilly E.M., Brody J.R., Witkiewicz A.K. Genetic diversity of pancreatic ductal adenocarcinoma and opportunities for precision medicine. Gastroenterology. 2016;150:48–63. doi: 10.1053/j.gastro.2015.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morikawa M., Derynck R., Miyazono K. TGF-beta and the TGF-beta family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 22.Principe D.R., Doll J.A., Bauer J., Jung B., Munshi H.G., Bartholin L., Pasche B., Lee C., Grippo P.J. TGF-beta: duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst. 2014;106:djt369. doi: 10.1093/jnci/djt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartholin L., Vincent D.F., Valcourt U. TGF-β as tumor suppressor: in vitro mechanistic aspects of growth inhibition. In: Moustakas A., Miyazawa K., editors. TGF-β in human disease. Springer Japan; Tokyo: 2013. pp. 113–138. [Google Scholar]
- 24.Pickup M., Novitskiy S., Moses H.L. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padua D., Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 26.Ikushima H., Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 27.Bierie B., Moses H.L. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Vincent D.F., Kaniewski B., Powers S.E., Havenar-Daughton C., Marie J.C., Wotton D., Bartholin L. A rapid strategy to detect the recombined allele in LSL-TbetaRI(CA) transgenic mice. Genesis. 2010;48:559–562. doi: 10.1002/dvg.20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartholin L., Cyprian F.S., Vincent D., Garcia C.N., Martel S., Horvat B., Berthet C., Goddard-Léon S., Treilleux I., Rimokh R., Marie J.C. Generation of mice with conditionally activated transforming growth factor beta signaling through the TβRI/ALK5 receptor. Genesis. 2008;46:724–731. doi: 10.1002/dvg.20425. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz A.L., Soudja S.M., Deceneux C., Lauvau G., Marie J.C. NK1.1+ CD8+ T cells escape TGF-beta control and contribute to early microbial pathogen response. Nat Commun. 2014;5:5150. doi: 10.1038/ncomms6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarron M.J., Marie J.C. TGF-beta prevents T follicular helper cell accumulation and B cell autoreactivity. J Clin Invest. 2014;124:4375–4386. doi: 10.1172/JCI76179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Havenar-Daughton C., Li S., Benlagha K., Marie J.C. Development and function of murine RORgammat+ iNKT cells are under TGF-beta signaling control. Blood. 2012;119:3486–3494. doi: 10.1182/blood-2012-01-401604. [DOI] [PubMed] [Google Scholar]
- 33.Viel S., Marcais A., Guimaraes F.S., Loftus R., Rabilloud J., Grau M., Degouve S., Djebali S., Sanlaville A., Charrier E., Bienvenu J., Marie J.C., Caux C., Marvel J., Town L., Huntington N.D., Bartholin L., Finlay D., Smyth M.J., Walzer T. TGF-beta inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal. 2016;9:ra19. doi: 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 34.Viant C., Rankin L.C., Girard-Madoux M.J., Seillet C., Shi W., Smyth M.J., Bartholin L., Walzer T., Huntington N.D., Vivier E., Belz G.T. Transforming growth factor-beta and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci Signal. 2016;9:ra46. doi: 10.1126/scisignal.aaf2176. [DOI] [PubMed] [Google Scholar]
- 35.Mohammed J., Beura L.K., Bobr A., Astry B., Chicoine B., Kashem S.W., Welty N.E., Igyarto B.Z., Wijeyesinghe S., Thompson E.A., Matte C., Bartholin L., Kaplan A., Sheppard D., Bridges A.G., Shlomchik W.D., Masopust D., Kaplan D.H. Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-beta. Nat Immunol. 2016;17:414–421. doi: 10.1038/ni.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y., Vincent D.F., Davis A.J., Sansom O.J., Bartholin L., Li Q. Constitutively active transforming growth factor beta receptor 1 in the mouse ovary promotes tumorigenesis. Oncotarget. 2016 doi: 10.18632/oncotarget.10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y., Duran S., Lydon J.P., DeMayo F.J., Burghardt R.C., Bayless K.J., Bartholin L., Li Q. Constitutive activation of transforming growth factor Beta receptor 1 in the mouse uterus impairs uterine morphology and function. Biol Reprod. 2015;92:34. doi: 10.1095/biolreprod.114.125146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadjantonakis A.K., Cox L.L., Tam P.P., Nagy A. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- 39.Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 41.Hameyer D., Loonstra A., Eshkind L., Schmitt S., Antunes C., Groen A., Bindels E., Jonkers J., Krimpenfort P., Meuwissen R., Rijswijk L., Bex A., Berns A., Bockamp E. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007;31:32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 42.Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vincent D.F., Yan K.P., Treilleux I., Gay F., Arfi V., Kaniewski B., Marie J.C., Lepinasse F., Martel S., Goddard-Leon S., Iovanna J.L., Dubus P., Garcia S., Puisieux A., Rimokh R., Bardeesy N., Scoazec J.Y., Losson R., Bartholin L. Inactivation of TIF1gamma cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5:e1000575. doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent D.F., Gout J., Chuvin N., Arfi V., Pommier R.M., Bertolino P., Jonckheere N., Ripoche D., Kaniewski B., Martel S., Langlois J.B., Goddard-Leon S., Colombe A., Janier M., Van Seuningen I., Losson R., Valcourt U., Treilleux I., Dubus P., Bardeesy N., Bartholin L. Tif1gamma suppresses murine pancreatic tumoral transformation by a smad4-independent pathway. Am J Pathol. 2012;180:2214–2221. doi: 10.1016/j.ajpath.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longnecker D.S., Lilja H.S., French J., Kuhlmann E., Noll W. Transplantation of azaserine-induced carcinomas of pancreas in rats. Cancer Lett. 1979;7:197–202. doi: 10.1016/s0304-3835(79)80080-4. [DOI] [PubMed] [Google Scholar]
- 46.Jessop N.W., Hay R.J. Characteristics of two rat pancreatic exocrine cell lines derived from transplantable tumors. Vitro. 1980;16:212. [Google Scholar]
- 47.Heldin C.-H., Landström M., Moustakas A. Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial–mesenchymal transition. Current Opinion in Cell Biology. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Ramjaun A.R., Tomlinson S., Eddaoudi A., Downward J. Upregulation of two BH3-only proteins, Bmf and Bim, during TGF beta-induced apoptosis. Oncogene. 2007;26:970–981. doi: 10.1038/sj.onc.1209852. [DOI] [PubMed] [Google Scholar]
- 49.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., Kawaguchi Y., Johann D., Liotta L.A., Crawford H.C., Putt M.E., Jacks T., Wright C.V., Hruban R.H., Lowy A.M., Tuveson D.A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhu L., Shi G., Schmidt C.M., Hruban R.H., Konieczny S.F. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gout J., Pommier R.M., Vincent D.F., Ripoche D., Goddard-Leon S., Colombe A., Treilleux I., Valcourt U., Tomasini R., Dufresne M., Bertolino P., Bartholin L. The conditional expression of KRAS G12D in mouse pancreas induces disorganization of endocrine islets prior the onset of ductal pre-cancerous lesions. Pancreatology. 2013;13:191–195. doi: 10.1016/j.pan.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Louhimo J., Steer M.L., Perides G. Necroptosis is an important severity determinant and potential therapeutic target in experimental severe pancreatitis. Cell Mol Gastroenterol Hepatol. 2016;2:519–535. doi: 10.1016/j.jcmgh.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanvito F., Herrera P.L., Huarte J., Nichols A., Montesano R., Orci L., Vassalli J.D. TGF-beta 1 influences the relative development of the exocrine and endocrine pancreas in vitro. Development. 1994;120:3451–3462. doi: 10.1242/dev.120.12.3451. [DOI] [PubMed] [Google Scholar]
- 54.Bottinger E.P., Jakubczak J.L., Roberts I.S., Mumy M., Hemmati P., Bagnall K., Merlino G., Wakefield L.M. Expression of a dominant-negative mutant TGF-beta type II receptor in transgenic mice reveals essential roles for TGF-beta in regulation of growth and differentiation in the exocrine pancreas. Embo J. 1997;16:2621–2633. doi: 10.1093/emboj/16.10.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J., Akanuma N., Liu C., Naji A., Halff G.A., Washburn W.K., Sun L., Wang P. TGF-beta1 promotes acinar to ductal metaplasia of human pancreatic acinar cells. Sci Rep. 2016;6:30904. doi: 10.1038/srep30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kopp J.L., von Figura G., Mayes E., Liu F.F., Dubois C.L., Morris JPt, Pan F.C., Akiyama H., Wright C.V., Jensen K., Hebrok M., Sander M. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duell E.J., Lucenteforte E., Olson S.H., Bracci P.M., Li D., Risch H.A., Silverman D.T., Ji B.T., Gallinger S., Holly E.A., Fontham E.H., Maisonneuve P., Bueno-de-Mesquita H.B., Ghadirian P., Kurtz R.C., Ludwig E., Yu H., Lowenfels A.B., Seminara D., Petersen G.M., La Vecchia C., Boffetta P. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4) Ann Oncol. 2012;23:2964–2970. doi: 10.1093/annonc/mds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagashio Y., Ueno H., Imamura M., Asaumi H., Watanabe S., Yamaguchi T., Taguchi M., Tashiro M., Otsuki M. Inhibition of transforming growth factor beta decreases pancreatic fibrosis and protects the pancreas against chronic injury in mice. Lab Invest. 2004;84:1610–1618. doi: 10.1038/labinvest.3700191. [DOI] [PubMed] [Google Scholar]
- 59.Yoo B.M., Yeo M., Oh T.Y., Choi J.H., Kim W.W., Kim J.H., Cho S.W., Kim S.J., Hahm K.B. Amelioration of pancreatic fibrosis in mice with defective TGF-beta signaling. Pancreas. 2005;30:e71–e79. doi: 10.1097/01.mpa.0000157388.54016.0a. [DOI] [PubMed] [Google Scholar]
- 60.Wildi S., Kleeff J., Mayerle J., Zimmermann A., Bottinger E.P., Wakefield L., Buchler M.W., Friess H., Korc M. Suppression of transforming growth factor beta signalling aborts caerulein induced pancreatitis and eliminates restricted stimulation at high caerulein concentrations. Gut. 2007;56:685–692. doi: 10.1136/gut.2006.105833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He J., Sun X., Qian K.Q., Liu X., Wang Z., Chen Y. Protection of cerulein-induced pancreatic fibrosis by pancreas-specific expression of Smad7. Biochim Biophys Acta. 2009;1792:56–60. doi: 10.1016/j.bbadis.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 62.Zion O., Genin O., Kawada N., Yoshizato K., Roffe S., Nagler A., Iovanna J.L., Halevy O., Pines M. Inhibition of transforming growth factor beta signaling by halofuginone as a modality for pancreas fibrosis prevention. Pancreas. 2009;38:427–435. doi: 10.1097/MPA.0b013e3181967670. [DOI] [PubMed] [Google Scholar]
- 63.Morris JPt, Wang S.C., Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.David C.J., Huang Y.H., Chen M., Su J., Zou Y., Bardeesy N., Iacobuzio-Donahue C.A., Massague J. TGF-beta tumor suppression through a lethal EMT. Cell. 2016;164:1015–1030. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oshimori N., Fuchs E. The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell. 2012;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massague J., Xi Q. TGF-beta control of stem cell differentiation genes. FEBS Lett. 2012;586:1953–1958. doi: 10.1016/j.febslet.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bardeesy N., Cheng K.H., Berger J.H., Chu G.C., Pahler J., Olson P., Hezel A.F., Horner J., Lauwers G.Y., Hanahan D., DePinho R.A. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Izeradjene K., Combs C., Best M., Gopinathan A., Wagner A., Grady W.M., Deng C.X., Hruban R.H., Adsay N.V., Tuveson D.A., Hingorani S.R. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 69.Kojima K., Vickers S.M., Adsay N.V., Jhala N.C., Kim H.G., Schoeb T.R., Grizzle W.E., Klug C.A. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res. 2007;67:8121–8130. doi: 10.1158/0008-5472.CAN-06-4167. [DOI] [PubMed] [Google Scholar]
- 70.Whittle M.C., Izeradjene K., Rani P.G., Feng L., Carlson M.A., DelGiorno K.E., Wood L.D., Goggins M., Hruban R.H., Chang A.E., Calses P., Thorsen S.M., Hingorani S.R. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell. 2015;161:1345–1360. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ijichi H., Chytil A., Gorska A.E., Aakre M.E., Fujitani Y., Fujitani S., Wright C.V., Moses H.L. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Principe D.R., DeCant B., Mascarinas E., Wayne E.A., Diaz A.M., Akagi N., Hwang R., Pasche B., Dawson D.W., Fang D., Bentrem D.J., Munshi H.G., Jung B., Grippo P.J. TGFbeta signaling in the pancreatic tumor microenvironment promotes fibrosis and immune evasion to facilitate tumorigenesis. Cancer Res. 2016;76:2525–2539. doi: 10.1158/0008-5472.CAN-15-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilentz R.E., Iacobuzio-Donahue C.A., Argani P., McCarthy D.M., Parsons J.L., Yeo C.J., Kern S.E., Hruban R.H. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- 74.Yachida S., Iacobuzio-Donahue C.A. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32:5253–5260. doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neuzillet C., de Gramont A., Tijeras-Raballand A., de Mestier L., Cros J., Faivre S., Raymond E. Perspectives of TGF-beta inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5:78–94. doi: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breitman T.R., Selonick S.E., Collins S.J. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci U S A. 1980;77:2936–2940. doi: 10.1073/pnas.77.5.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Breitman T.R., Collins S.J., Keene B.R. Terminal differentiation of human promyelocytic leukemic cells in primary culture in response to retinoic acid. Blood. 1981;57:1000–1004. [PubMed] [Google Scholar]
- 78.Fitamant J., Kottakis F., Benhamouche S., Tian H.S., Chuvin N., Parachoniak C.A., Nagle J.M., Perera R.M., Lapouge M., Deshpande V., Zhu A.X., Lai A., Min B., Hoshida Y., Avruch J., Sia D., Camprecios G., McClatchey A.I., Llovet J.M., Morrissey D., Raj L., Bardeesy N. YAP inhibition restores hepatocyte differentiation in advanced HCC, leading to tumor regression. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]