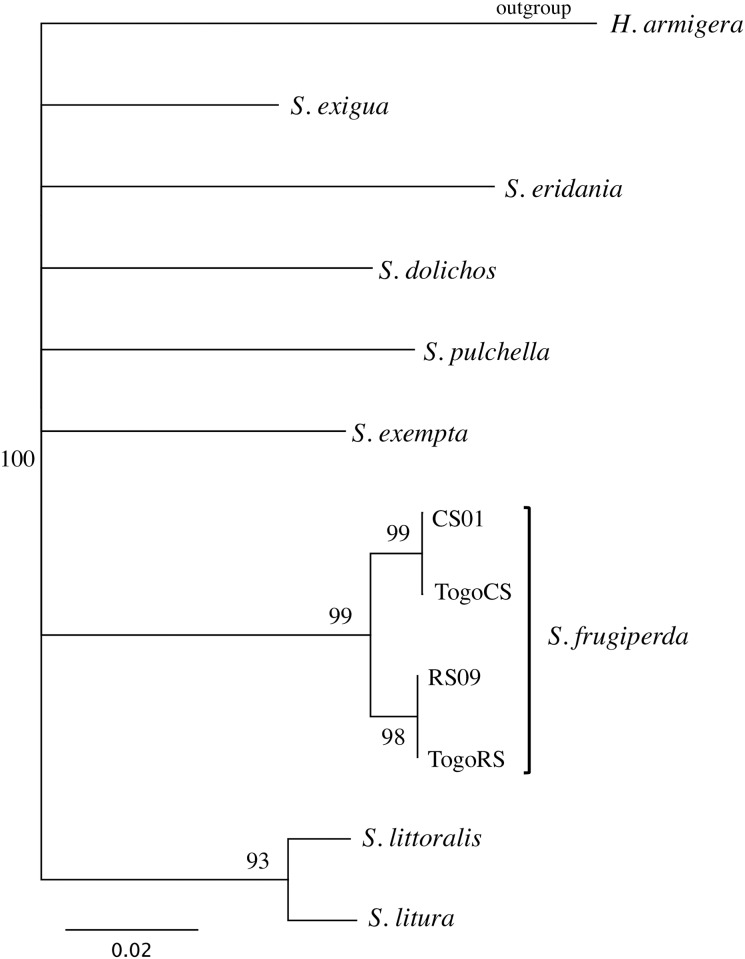
Fig 3. Strict consensus phylogenetic tree derived from neighbor- joining analysis comparing the two Togo barcode sequences (Togo CS, Togo RS) with those from fall armyworm host strains and related Spodoptera species [35].
The sequences from the Spodoptera species are the consensus of the following variants found in Genbank, S. dolichos (HM756086-9), S. eridania (HM756081-5), S. exempta (HQ177331-6, DQ092371-6), S. exigua (HM756077-80), S. litura (HM6090-3), S. littoralis (HM756074), S. pulchella (756075–6). The two distinct Togo sequences were identical to the fall armyworm corn strain haplotype CSO1 (HM136586) and rice strain haplotype RS01 (HM136601). The phylogenetic analysis was based on a 402-bp segment of the CO1 gene common to all sequences with the equivalent CO1 segment from Helicoverpa armigera (KM275101) used as the outgroup. The tree is based on Kimura-2-Parameter distances. Numbers at branch points indicate 2000X bootstrap values. Scale bar represents substitutions per site.