Figure 1. Genomic annotation of the 3p22.1, 7p15.3, and 22q13.1 regions.
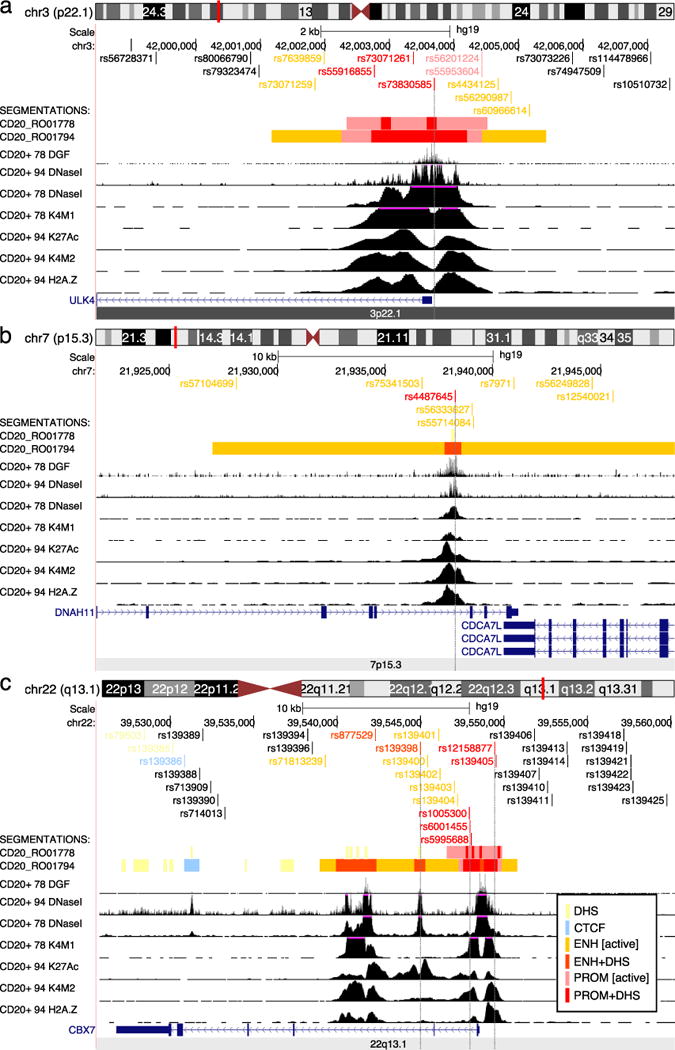
UCSC browser views showing wiggle tracks from ENCODE data for CD20+ B-cells from two cell lines, RO01778 and RO01794. The peak calls from these data were used to segment the genome into non-coding functional regions as detailed in the inset at bottom right. a) Region 3p22.1 detailing the 5′ end of the ULK4 gene, where high-confidence SNPs overlap the central regulatory core region of the active promoter. b) Overview of the 7p15.13; an enhancer with active histone marks within intron 79 of DNAH11 as described in the text. c) Overview of region 22q13.1 where several SNPs overlap with the promoter and downstream enhancers of CBX7. DHS: DNase I hypersensitive site, CTCF: CTCF bound region, ENH [active]: enhancer with H3K27 acetylation (K27Ac), ENH+DHS: DNase I hypersensitive region found within an active enhancer, PROM [active]: promoter with H3K27 acetylation, PROM+DHS: DNase I hypersensitive region found within an active promoter. Other abbreviations: DGF: digital DNase I footprinting, K4M1: H3K4 mono-methylation, K4M2: H3K4 dimethylation, H2A.Z: H2A.Z histone modification (not used for segmentations).
