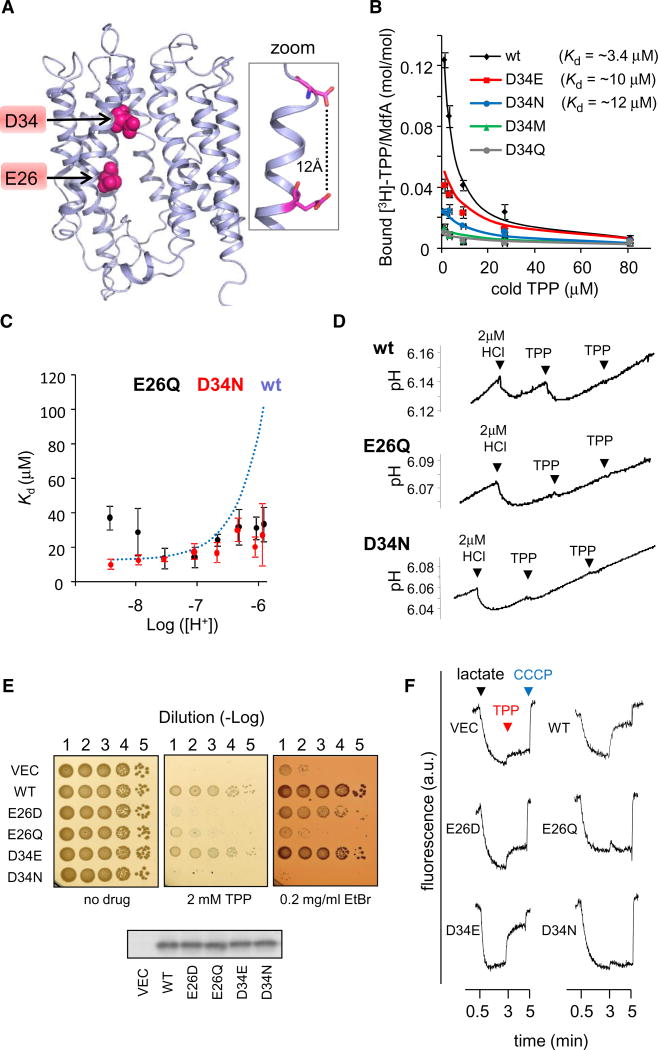
Figure 3. Characterization of E26 and D34 Mutants.
(A) Location of E26 and D34 in the 3D-structural model of MdfA. The 11th transmembrane segment is not shown for clarity. The inset on the right shows the calculated distance between E26 and D34.
(B) Inhibition by cold TPP of [3H]-TPP binding to the various D34X mutants. Dissociation constants obtained from nonlinear regression fitting (solid line) are indicated. Error bars indicate SD.
(C) [H+]-dependence of the affinity of MdfA(E26Q) or MdfA(D34N) for TPP, as determined by PEGylation protection in membrane vesicles. The dependence of the wild-type, as determined in Figure 1E is shown for comparison (dotted line). Error bars indicate SD.
(D) Time-dependent pH measurements of proton release by MdfA(E26Q) and MdfA(D34N) (see Figure 2B).
(E) Drug-resistance of E. coli cells harboring the indicated MdfA mutant (or empty vector). Serial dilutions of cells were spotted on drug-supplemented media. Western blotting analysis shows that the expression of the various MdfA mutants is comparable (lower panel).
(F) Proton transport by everted membrane vesicles containing different MdfA mutants. The trans-membrane ΔpH was followed by measuring ACMA fluorescence. Lactate (added after 0.5 min) and CCCP (after 5 min) were used to generate or dissipate ΔpH, respectively. TPP was added after 3 min.