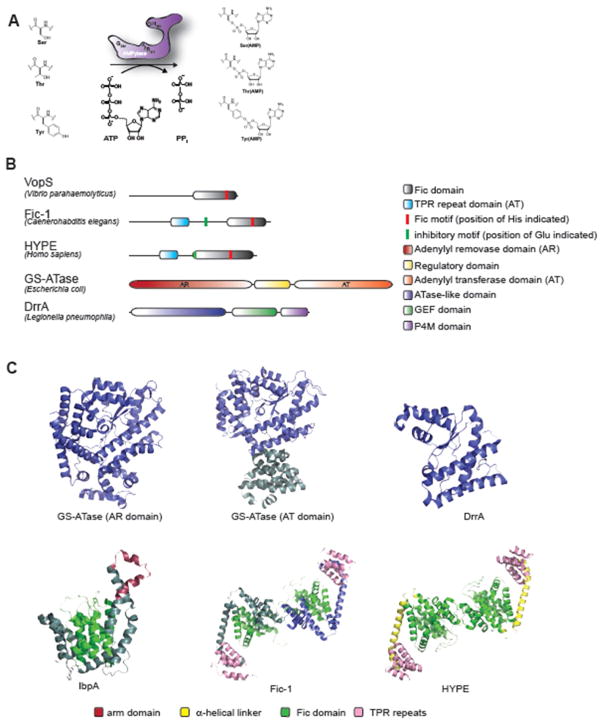
Figure 1. Organization and structure of prokaryotic and eukaryotic AMPylases.
(A) Schematic representation of AMP transfer to Serine (Ser), Threonine (Thr) and Tyrosine (Tyr) catalyzed by a Fic-domain containing AMPylase. (B) Proportional representation of domain organization of fic-domain containing AMPylases VopS (V. parahaemolyticus), Fic-1 (C. elegans) and HYPE (H. sapiens) as well as non-fic AMPylases DrrA (L. pneumophila) and GS-ATase (E. coli). Key amino acids of the active site motif (His) and inhibitory motif (Glu) are highlighted in red and green, respectively. (C) Structures of five representative AMPylases. GS-ATase (AR domain PDB ID: 1V4A; AT domain PDB ID: 3K7D) (Xu et al., 2010, Xu et al., 2004), DrrA (PDB ID: 3NKU) (Müller et al., 2010) (both non-fic) as well as IbpA (PDB: 3N3U) (Xiao et al., 2010) are presented as monomers. Fic-1 (PDB ID: 5JJ7) (Truttmann et al., 2016) and HYPE (PDB ID: 4U07) (Bunney et al., 2014) are shown as inverted homo-dimers (as found in solution). Dimerization is mediated exclusively through interactions between the two fic domains while TPR domains do not participate and extend from the individual monomers.