Figure 2.
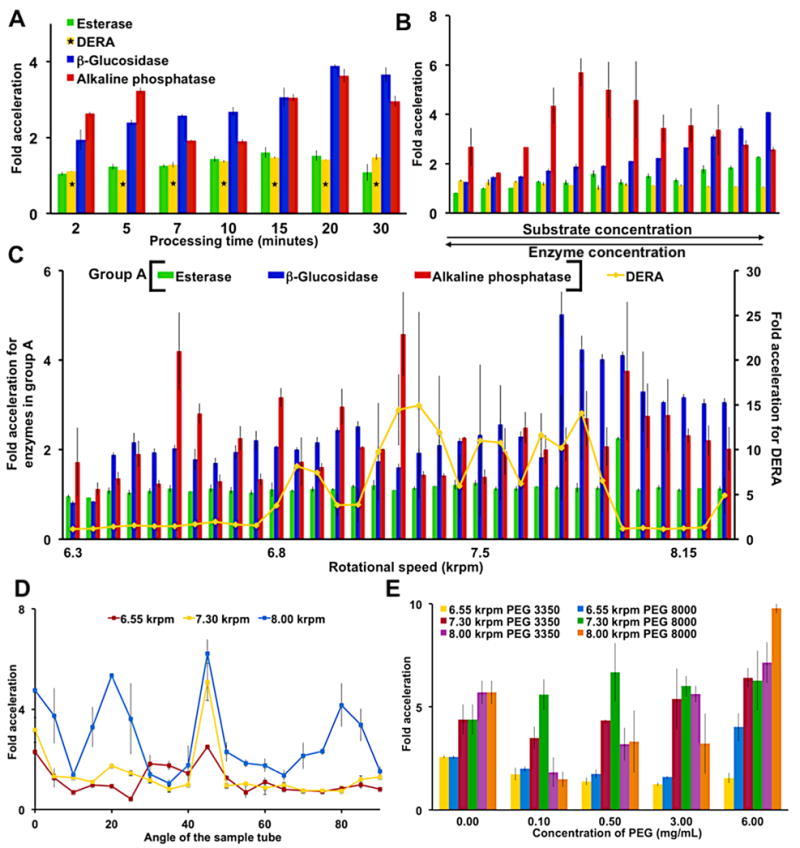
Parameters for accelerated biocatalysis of the four enzymes. Fold acceleration was determined by the ratio of the VFD-mediated substrate conversion to an identical enzyme-substrate solution not treated by the VFD. (A) A time dependent study at a fixed rotational speed (8.00 krpm) reveals processing times for further optimization. As indicated with a DERA required longer reaction times of 60, 80, 100, 120, 140, 160 and 180 min. (B) Simultaneous changes to the substrate and enzyme concentrations at a 8.00 krpm rotational speed mapped the reaction landscape (Table S2A–D). (C) Rotational speed scans in 50 rpm increments identify harmonic oscillations associated with Faraday wave-promoted biocatalysis. Error bars are larger for DERA than any other enzyme due to the product’s non-linear, fluorescence calibration curve. (D) Varying the tilt angle of the sample tube identifies 45° as optimal. Thus, a 45° tilt angle was used throughout this report. (E) The addition of PEG dramatically slowed the non-VFD control, but the VFD processed solution demonstrated significant catalytic activity. Error bars indicate the standard deviation around the mean (n=3 with three independent measurements on three different VFDs). With the exception of a single data point requiring 90% confidence limits, all data reported here have no overlapping errors within 95% confidence limits. The concentrations of enzymes and substrates are as follows: alkaline phosphatase (6.77 nM) and its substrate p-nitrophenol phosphate (0.17 mM), β-glucosidase (19.3 nM) and its substrate 4-nitrophenyl β-D-glucopyranoside (7.5 mM), esterase (0.12 nM) and its substrate p-nitrophenol acetate (44 μM) and DERA (7.69 μM) and its fluorogenic substrate (0.52 mM) unless otherwise indicated, and as described in Table S2A-D.
