Fig. 2.
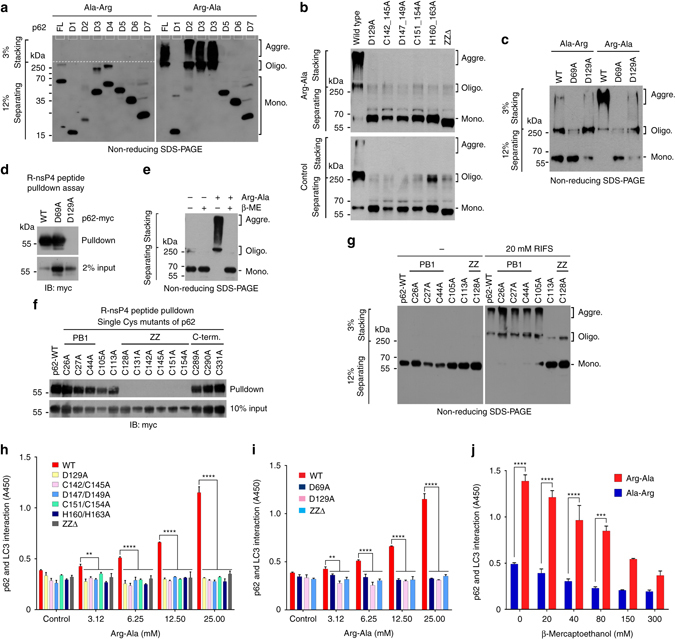
The binding of Nt-Arg to p62 ZZ domain facilitates disulfide bond-linked aggregation of p62 and p62–LC3 interaction. a In vitro oligomerization assay using myc/His tagged p62 deletion mutants (Supplementary Fig. 2a), followed by non-reducing SDS-PAGE and immunoblotting using a mixture of antibodies to p62 and Myc. b In vitro oligomerization assay using myc/His tagged wild type p62 and mutants carrying point mutations in ZZ domain. c In vitro p62 oligomerization assay using p62 D69A and D129A mutants in comparison with wild-type p62. d R-nsP4 pulldown assay using constructs used in c. e In vitro oligomerization assay of p62 ectopically expressed in HEK293 cells treated with 50 mM Arg-Ala in the presence of 50 mM β-mercaptoethanol (β-ME). f R-nsP4 peptide binding assay of p62 Cys mutants. Each Cys/Ala p62 mutant with myc/His tag was expressed in HEK293 cells, and 50 μg of total protein was used for pulldown. g In vitro oligomerization assay of p62 Cys mutants used in f. Cell lysates with ectopically expressed wild-type p62 and Cys mutants that can bind Nt-Arg were incubated with 20 mM RIFS tetrapeptide for 1.5 h at room temperature. h ELISA measuring the interaction of p62 ZZ domain mutants with LC3. Cell lysates overexpressing p62 proteins were incubated with Arg-Ala at different concentrations to allow p62 binding to LC3-GST linked to glutathione coated plates. Shown are the means (±S.D.) of three independent experiments, each performed in triplicate. A one-way ANOVA was performed to determine statistical significance (**P < 0.01; ****P < 0.0001). i ELISA measuring p62D69A interaction with LC3 as described in h. The graph represents the mean (±S.D.) of three independent experiments. Statistical significance was calculated using a one-way ANOVA test (**P < 0.01; ****P < 0.0001). j Similar to i except that p62–LC3 interaction was measured in the presence of 25 mM Arg-Ala or Ala-Arg in combination with β-mercaptoethanol (β-ME) at concentrations indicated. Data represent the mean (±S.D.) of three independent experiments. Statistical significance was determined using a one-way ANOVA test (***P < 0.001; ****P < 0.0001)
