Figure 1.
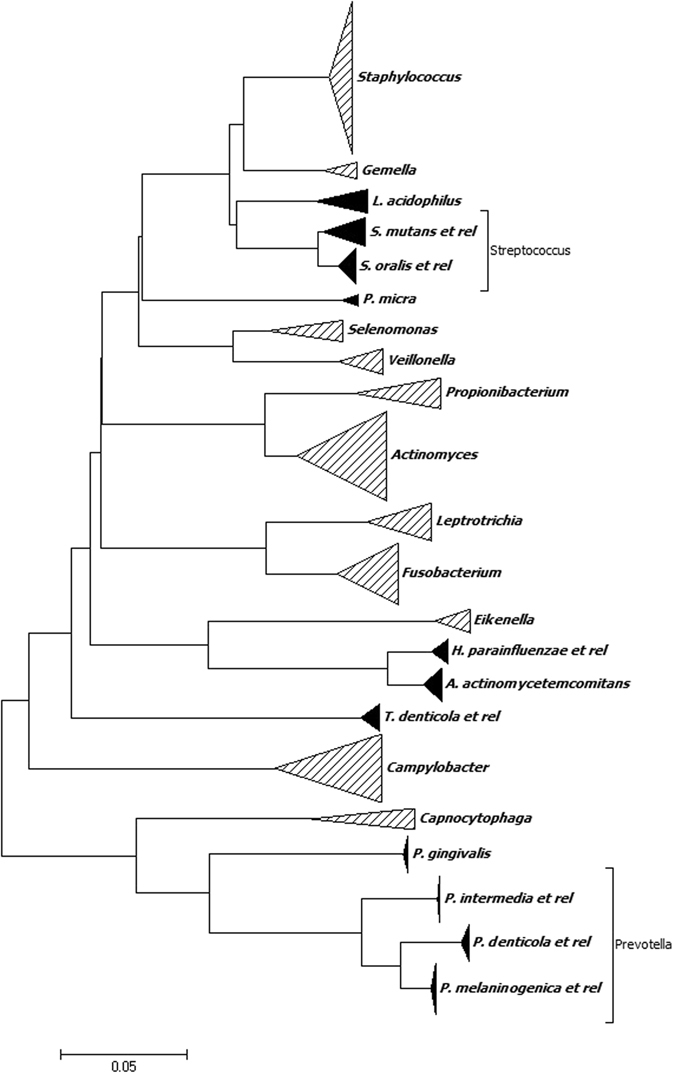
Phylogenetic tree of the oral microrganisms targeted by the OralArray. The tree was obtained from the entire positive set used for the probe design. The neighbor-joining method was used to infer evolutionary history. The evolutionary distances were computed using the maximum composite likelihood method and units correspond to the number of base substitutions per site. The analysis involved 383 nucleotide sequences (Supplementary Table S2). All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted by using MEGA6 software.
