Figure 2.
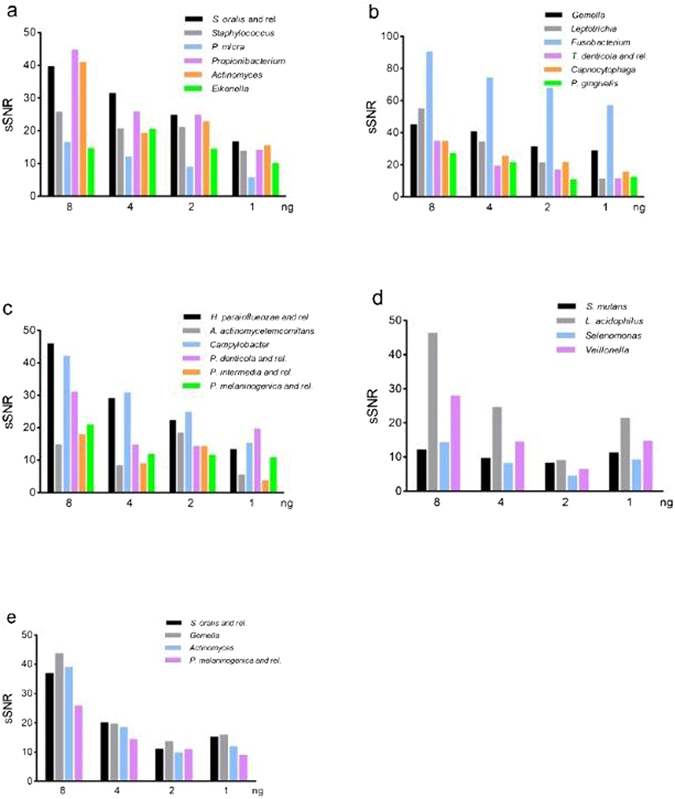
Sensitivity of the OralArray on DNA mixes. Artificial mixes of 16 S rRNA amplicons were subjected to LDR-UA analysis, sSNR values were plotted vs ng of each DNA amplicon. sSNR values were obtained with decreasing amounts of DNA mixes containing: (a) S. oralis, S. aureus, P. micra, P. acidifaciens, A. odontolyticus, and E. corrodens 16 S rRNA amplicons (8–1 ng each); (b) G. morbillorum, L. buccalis, F. nucleatum, T. denticola, C. sputigena, and P. gingivalis 16 S rRNA amplicons (8–1 ng each); (c) H. parainfluenzae, A. actinomycetemcomitans, C. sputorum, P. denticola, P. intermedia, and P. melaninogenica 16 S rRNA amplicons (8–1 ng each); (d) S. mutans, L. acidophilus, S. noxia, and V. parvula 16 S rRNA amplicons (8–1 ng each); (e) S. oralis, G. morbillorum, A. odontolyticus, and P. melaninogenica 16 S rRNA amplicons (8–1 ng each) added with human genomic DNA up to 100 ng of total DNA (68–96 ng).
