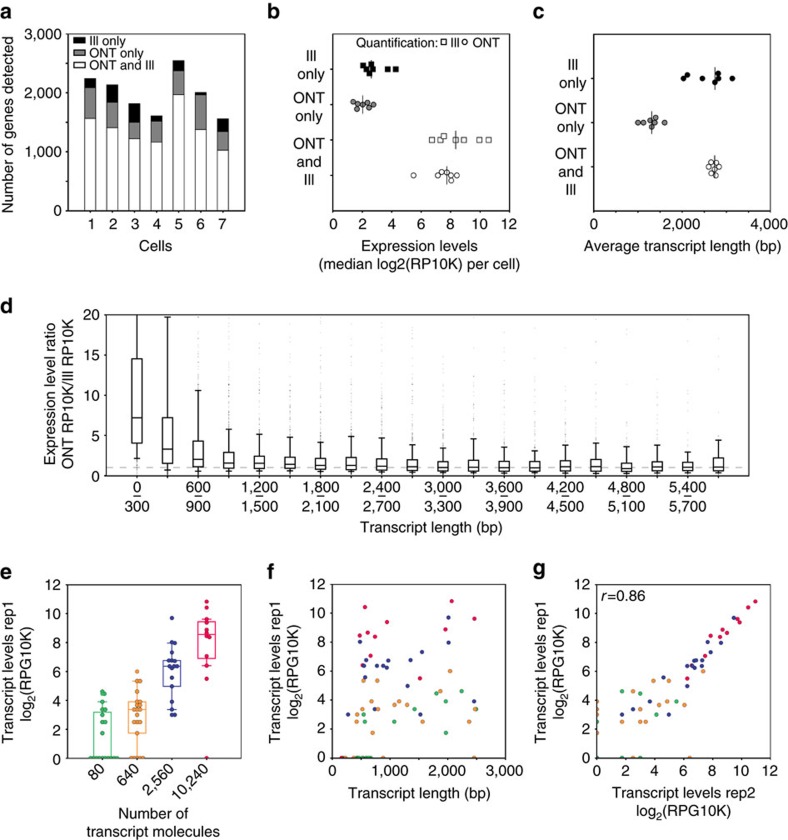
Figure 3. Quantifying gene and transcript expression with ONT RNAseq data.
(a) Stack barplots showing the number of genes detected in each cell corresponding to different sequencing technologies (Ill—Illumina, ONT—Oxford Nanopore). (b) Median expression levels of genes detected by both or individual technologies. Two expression levels (Ill and ONT) are given for genes detected in both technologies. (c) Gene length of genes detected by both or individual technologies. (d) Ratio of gene expression levels for genes detected by both technologies. Ratios are binned according to gene length and shown as boxplots with whiskers indicating 10th and 90th percentiles. (e) SIRV transcript levels of Replicate 1 (Rep1: 100fg SIRV pool E2) as measured with ONT RNAseq. Transcripts are binned by their starting molecule numbers. (f) SIRV transcript levels of Replicate 1 are plotted against transcript length with colours corresponding to groups in e. (g) Scatter plot showing correlation of SIRV transcript expression levels of Replicate 1 (Rep1: 100fg SIRV pool E2) and Replicate 2 (Rep2: 100 fg SIRV pool E2), both measured by ONT RNAseq. r-value shown is Pearson r. Colours corresponding to groups in e.