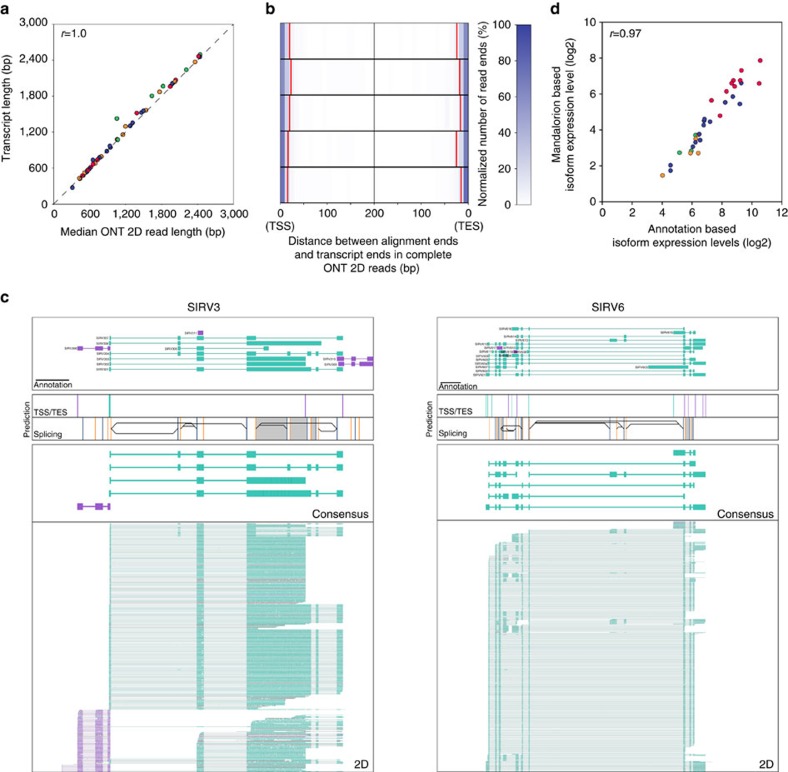
Figure 4. Identifying and quantifying transcript isoforms in SIRV E2 mixtures.
(a) Scatter plot shows correlation between ONT 2D reads and the SIRV transcripts they align to. Pearson r is shown. Colouring is as indicated in Fig. 3e–g (b) Distance between read alignment ends and transcript ends are shown as heatmap with the colour indicating the normalized alignment numbers. 90% of read alignments terminated outside the red lines (c) Genome Browser view of SIRV3(c) and SIRV6(d) gene loci. Top box contains transcript annotations with black 1 kb scale bar in bottom left corner, second and third box contain TSS (Teal)/TES (Purple) and splice sites (yellow: 5′SS, blue: 3′SS) locations predicted from the read data, respectively. Black lines and grey areas in box 3 indicate alternative splicing and intron retention events predicted from the read data. Box 4 contains read alignments of isoform consensus reads. Box 5 contains ONT 2D read alignments. Direction of transcripts, isoform consensus, and ONT 2D reads are indicated by their colour (Teal: 5′ to 3′, Purple: 3′ to 5′). (d) Scatter plot shows correlation between SIRV transcript quantification by aligning to annotated transcripts or by annotation-independent isoform grouping using Mandalorion. Pearson r is shown.