Figure 1. PmScsC function.
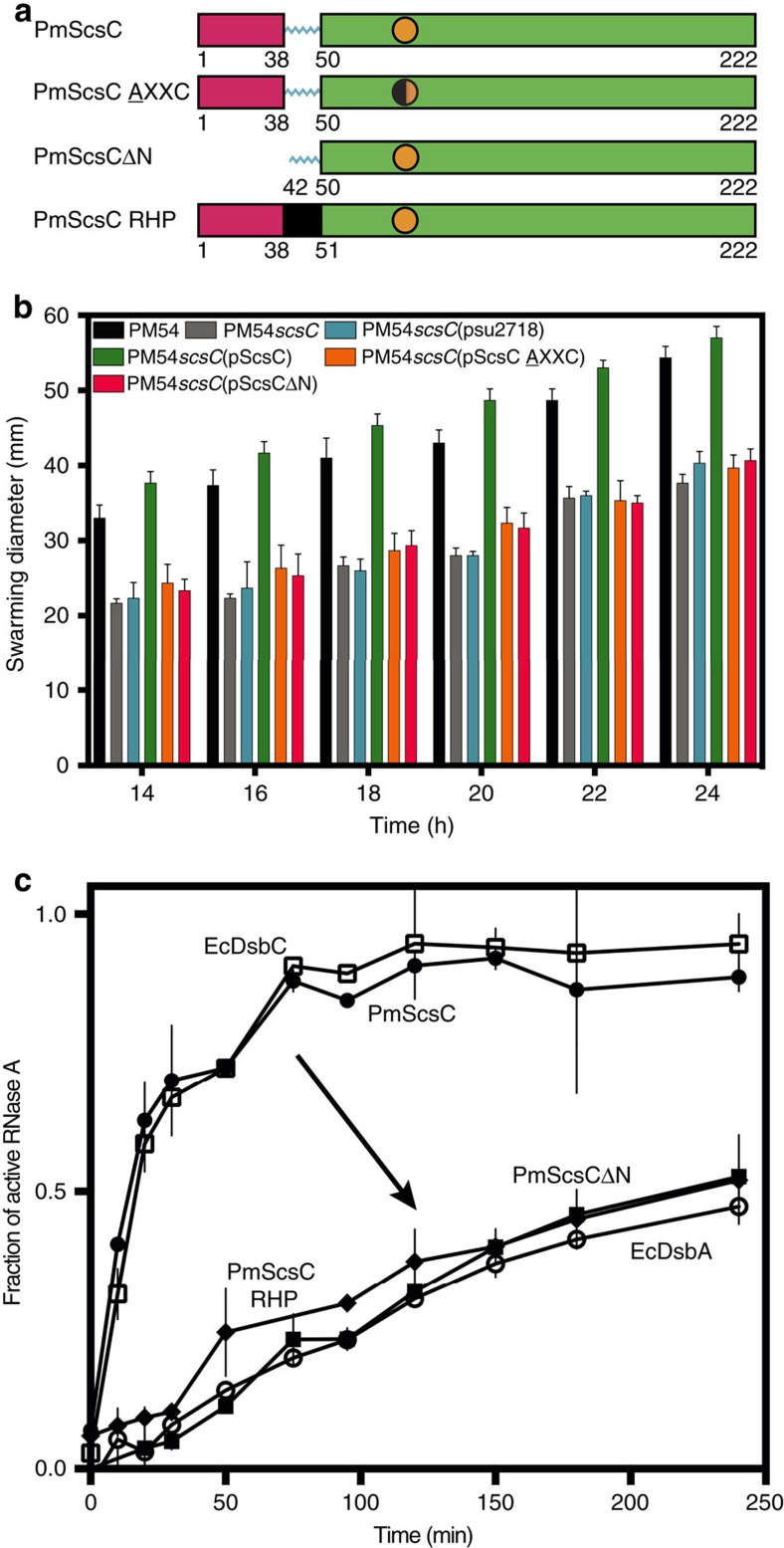
(a) Linear representation of the domain organization of PmScsC mutants. The trimerization domain is coloured magenta, the catalytic domain is green, with the CXXC motif represented as an orange circle, and the linker region is shown in cyan. The rigid helical linker is shown in black. (b) Swarming motility of wild type P. mirabilis strain PM54 (black), PmScsC deletion mutant PM54scsC (grey) and PM54scsC containing control and complementation plasmids: PM54scsC(pSU2718) (vector control; cyan), PM54scsC(pScsC) (wild-type; green), PM54scsC (pScsC AXXC) (active site mutant; orange) and PM54scsC(pScsCΔN) (N-terminal trimerization domain deletion; magenta) in the presence of 1.5 mM CuSO4 on LB agar plates (significant difference, P<0.0001 for slope calculated by F-test). Data are shown as the mean±s.d., of a single experiment performed in triplicate; all data is representative of three independent experiments. (c) Disulfide isomerase activity of PmScsC (hollow squares) is similar to that of EcDsbC (filled circles). Monomeric PmScsΔN (filled squares) and the PmScsC mutant engineered to have a rigid helical peptide (RHP) in place of the flexible peptide (filled diamonds) show negligible activity (equivalent to oxidase EcDsbA (hollow circles)). Data are shown as the mean±s.d., of two replicate experiments.